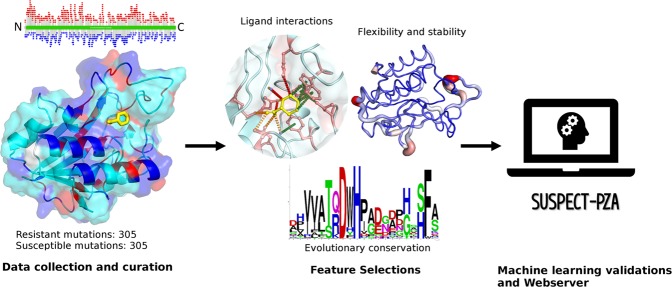
Figure 1.
Methodology workflow. The methodology can be divided into three steps. In step 1, data is collected and curated from various tuberculosis databases and articles with experimental evidence like availability of DST results or high-precision laboratory screening study. The curated mutations are shown across both the protein sequence and 3D structure, respectively. The protein sequence and structure of PncA is colored by whether resistant (red) or susceptible (blue) mutations have been observed at that location. Highlighting the difficulty of genomic analysis of pncA, both resistant and susceptible mutations have been observed across many residue positions (cyan). In step 2, effects of mutations on protein stability, dynamics, complementary information regarding the environment characteristics of the wild-type residue (e.g. relative solvent accessibility, residue depth and secondary structure), PZA binding affinity are calculated using different in-silico tools. Step 3, all the features are used as evidence to train a supervised machine learning algorithm and after evaluating the performance of the predictive model, the consensus predictions are integrated into a server and can be used to guide clinical resistance detection.