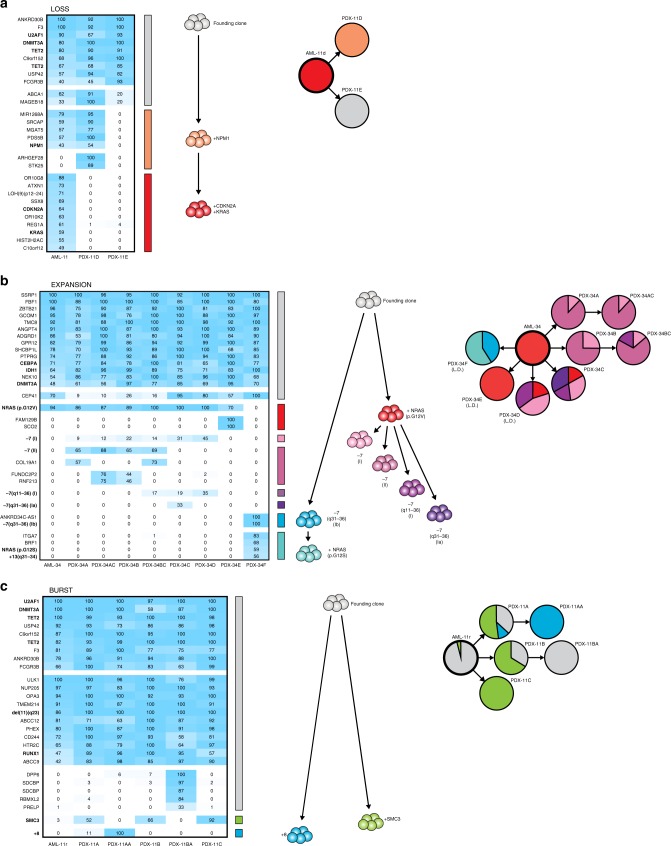
Fig. 3. The clonal composition changes in the majority of AML xenografts.
a A representative AML case with the Loss pattern of clonal dynamics, where a subclone in the patient sample is reduced or completely lost in the xenografts. In AML-11d, the dominant clone in the patient sample, with CDKN2A and KRAS mutations, was lost in both xenografts, resulting in engraftment with one of two parental clones. Left, the percentage of cells in patient samples and corresponding xenografts estimated to carry each genetic aberration, based on variant allele frequencies of identified mutations and b-allele frequencies of copy number alterations and copy-neutral losses of heterozygosity. Colored bars indicate defining mutations for each clone. Clones are represented by the same color throughout each panel. Middle, inferred clonal hierarchy. Right, proportions of each clone at diagnosis and in PDXs. Clones were defined by the presence of one or more recurrent AML mutations, CNAs or losses of heterozygosity (indicated in bold). L.D. denotes samples transplanted at minimal cell dose based on limiting dilution analysis. b A representative AML case with the Expansion pattern of clonal dynamics, where a subclone in the patient sample expands to constitute the entirety of the xenografts. In AML-34, clones with one of two different NRAS mutations and five different partial or complete losses of chromosome 7, which were all undetectable at diagnosis, expanded to generate the xenografts. The heterozygous CEP41 mutation is present in the founding clone but located on chromosome 7 and thus lost in the −7 (II) clone but retained in the −7 (I) clone. I and II denote the two alleles of chromosome 7, whereas a and b represent distinct genetic events. c A representative AML case with the Burst pattern of clonal dynamics, where a subclone in the patient sample expands in primary xenografts but is lost in secondary xenografts. In AML-11r, a small subclone with an SMC3 mutation transiently expanded to make up the majority of both primary recipients but was lost in the two secondary recipients. The +8 aberration was detected at very low frequency in the diagnostic sample by routine clinical karyotyping.