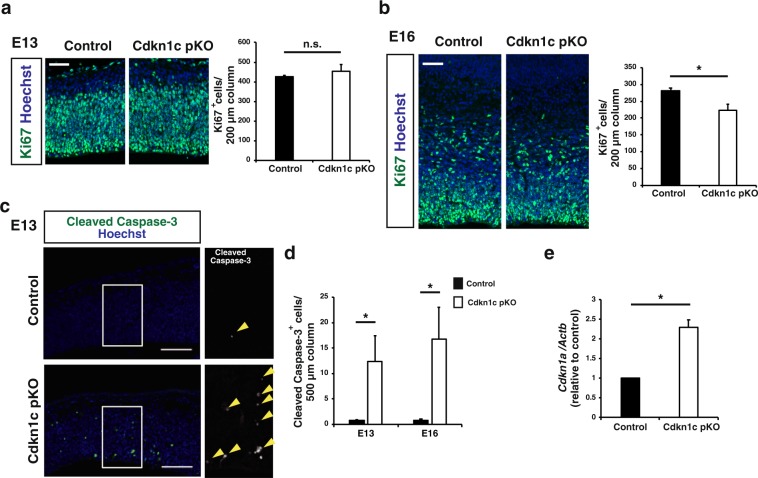
Figure 4.
CNS-specific deletion of the Cdkn1c paternal allele reduced the number of proliferating progenitors at E16 and increased the number of apoptotic cells at E13 and E16. (a,b) Immunofluorescence staining of Ki67 at E13 (a) and E16 (b) (left panel). Quantitative analysis of cells positive for Ki67 per area within 200 μm wide bins (right panel) (n = 3–4 embryos for each genotype). Data are mean + s.e.m. Unpaired two-tailed Student’s t-test. (c) Immunostaining of cleaved caspase-3 at E13. The boxed regions in the left panels are shown at a higher magnification in the right panel. Yellow arrowheads indicate cleaved caspase-3 positive cells. (d) The number of cleaved caspase-3 positive cells per area within 500 μm wide bins were quantified at E13 and E16 (n = 4–6 embryos for each genotype). Data are mean + s.e.m. Unpaired two-tailed Student’s t-test. (e) qPCR of Cdkn1a mRNA expression in the neocortex isolated from control and paternal Cdkn1c cKO mice at P0. Cdkn1a mRNA expression was normalized to β-actin (n = 3 independent experiments). Data are mean + s.e.m.; expressed relative to the corresponding value for control mice. Paired two-tailed Student’s t-test. *P < 0.05. n.s., not significant. Scale bars: 50 μm in (a,b); 100 μm in (c).