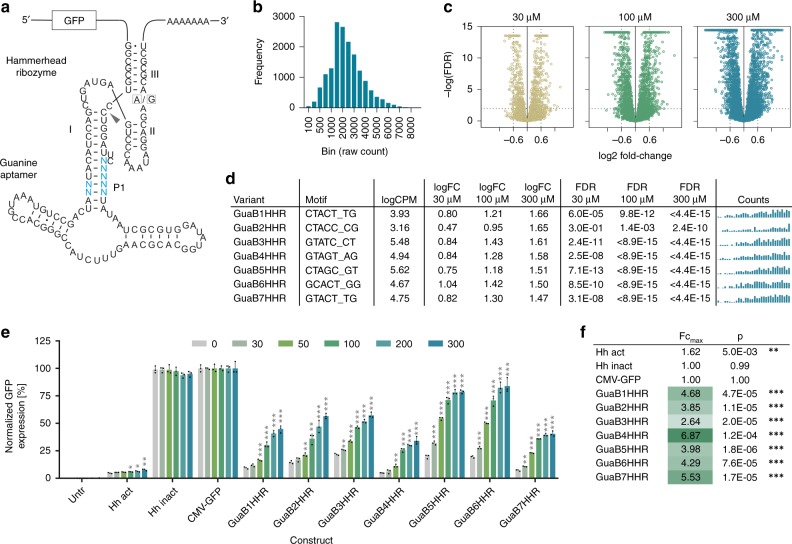
Fig. 4. Identification of Gua-responsive hammerhead ribozymes by amplicon-seq.
a Secondary structure of the guanine hammerhead ribozyme library design. Blue nucleotides indicate the randomized motif. The arrowhead indicates the cleavage site. A/G mutation at the indicated position renders the ribozyme inactive. b Guanine riboswitch construct frequency analysis of the original library. c Volcano plots displaying the significance (FDR) of changes in riboswitch abundance as a function of the change in expression (log2 fold change). d Selected hits from the Gua-HHR riboswitch library screen, randomized sequence, mean counts per million (CPM), fold change (FC) and false-discovery rate (FDR) at 30, 100, and 300 µM guanine, respectively. The bar plots show the raw counts under untreated, 30, 100, and 300 µM µM Gua-stimulated conditions (four groups from left to right, n = 8 replicates each). e Functional hit validation in HEK-293 cells at increasing guanine doses [µM] (n = 3 replicates, mean ± s.d.); Hh-act/inact: constitutively active/inactive ribozyme control. f Maximal fold changes and p-values measured upon stimulation vs. 0 µM guanine for all tested constructs. *p < 0.05, **p < 0.01, ***p < 0.001 (unpaired T-Test, two-tailed). Source data are provided as a Source Data file.