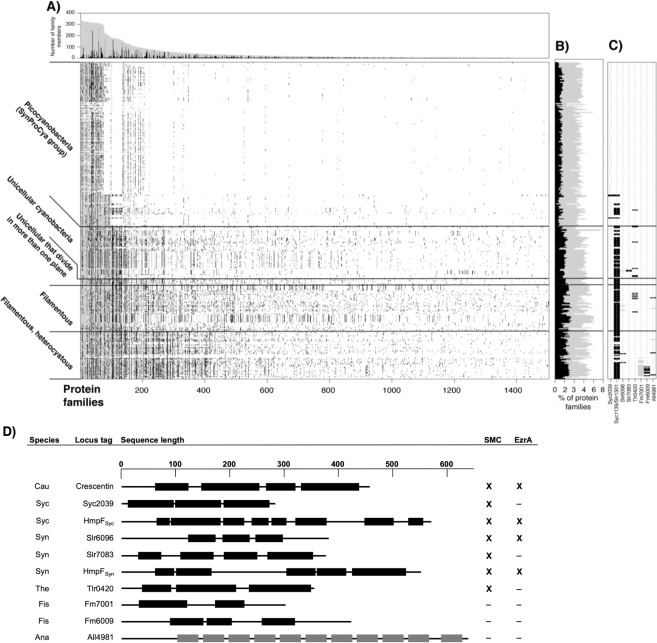
Figure 1.
Distribution of CCRP protein families within cyanobacteria. (A) Lines in the presence/absence matrix designate cyanobacterial genomes; each column shows a protein family. Gray dots designate any homologous protein in the same protein family and black dots represent CCRP members. Protein families are sorted according to the number of members. Protein family size and the number of CCRP members are presented in a bar graph above. (B) The proportion of protein families containing CCRPs (gray) and CCRP proteins (black) in each genome. (C) Presence/absence pattern of CCRP candidate protein families. Only protein families with at least three members predicted to be CCRPs are shown. (D) Domain prediction of CCRP candidates. Scale on top is given in amino acid residues. Amino acid sequences in coiled-coil conformation are depicted by black bars with non-coiled-coil sequences represented by black lines. Tetratricopeptide repeats (TPR), also predicted by the COILS algorithm, are shown as grey bars. Proteins are given as cyanobase locus tags. Fm7001 and Fm6009 correspond to NCBI accession numbers WP_016868005.1 and WP_020476706, respectively. Abbreviations: Cau: C. crescentus; Syc: Synechococcus, Syn: Synechocystis; Ana: Anabaena; The: Thermosynechococcus elongatus BP-1; Fis: Fischerella. Cyanobacterial CCRPs had conserved domains present in prokaryotic IF-like CCRPs and eukaryotic IF proteins (Supplementary Table 1). Presence of a structural maintenance of chromosomes (SMC) domain or structural similarities to the cell division protein EzrA are marked with “X”, absence is indicated with “−”. Full list is given in Supplementary Table 1.