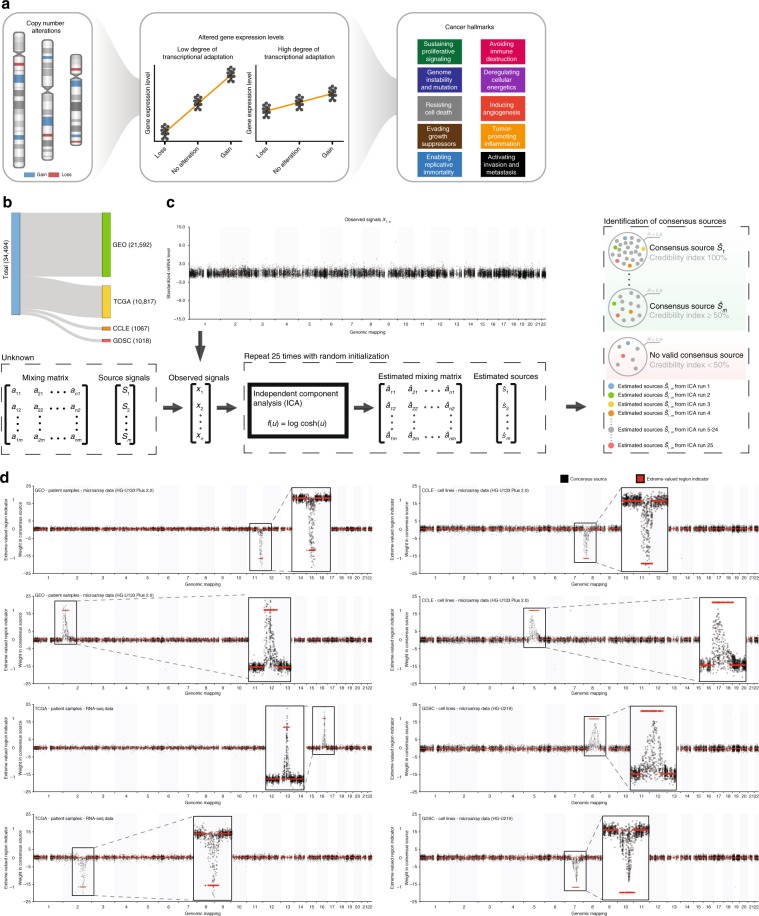
Fig. 1. Data acquisition and decomposition of gene expression profiles.
a CNAs can promote tumor progression via altering expression levels of genes located at the affected genomic regions. However, due to transcriptional adaptation mechanisms, changes in gene copy number at the genomic level do not always translate proportionally into altered mRNA expression levels. Unraveling the degree of transcriptional adaptation to CNAs for all genes will greatly contribute to our knowledge how CNAs drive tumor progression. b Number of gene expression profiles collected from GEO, TCGA, CCLE, and GDSC. c Identification of underlying regulatory factors of the mRNA transcriptome. We hypothesized that the observed gene expression in a gene expression profile is the result of (i) the effect of underlying regulatory factors (i.e., source signals) on expression levels of individual genes and (ii) the activity of these underlying regulatory factors in a complex biopsy (i.e., mixing matrix). ICA was used to capture the number and nature of these underling regulatory factors for all four datasets separately. This resulted in estimated sources, representing the effects of independent underlying regulatory factors on the expression levels of individual genes, and a mixing matrix reflecting the activity of each estimated source in each gene expression profile. ICA was run 25 times with random initialization, followed by consensus sources estimation using a credibility index ≥50%. d Examples of CNA-CESs harboring a pattern in which only genes mapping to a specific contiguous genomic region had a high absolute weight. The red line shows whether genomic regions were marked as having a significant number of genes with a high absolute weight by the detection algorithm (i.e., extreme-valued region indicator).