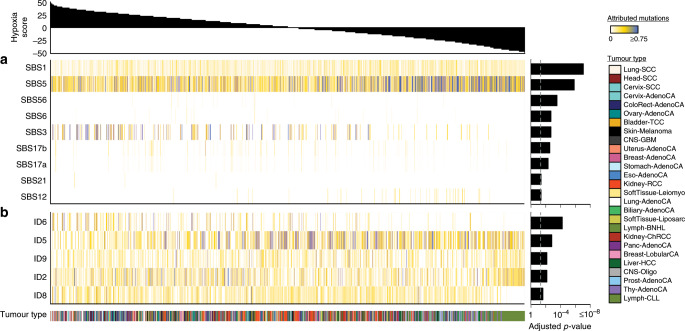
Fig. 3. Hypoxia-associated mutational signatures.
We associated hypoxia with the proportion of mutations attributed to specific mutational signatures using linear-mixed effect models. Hypoxia scores for 1188 independent tumours are shown across the top while FDR-adjusted p-values are shown on the right. a Hypoxia was associated with a series of single base substitution signatures with unknown aetiology including SBS5, SBS17a, SBS17b and SBS12. Some of these mutational signatures may reflect hypoxia-dependent mutational processes. Hypoxia was also associated with a lower proportion of attributed mutations to SBS1, which reflects deamination of 5-methylcytosine, and a higher proportion of attributed mutations to SBS3, which is related to deficiencies in DNA double-strand break repair and homologous recombination. b Several signatures of small insertions and deletions were also associated with hypoxia, including ID6 and ID2, which reflect defective homologous recombination and defective DNA mismatch repair, respectively. ID5, ID9 and ID4, all with unknown aetiology, were significantly associated with hypoxia score. All associations were modelled using linear-mixed effect models while adjusting for cancer type, tumour purity, age and sex.