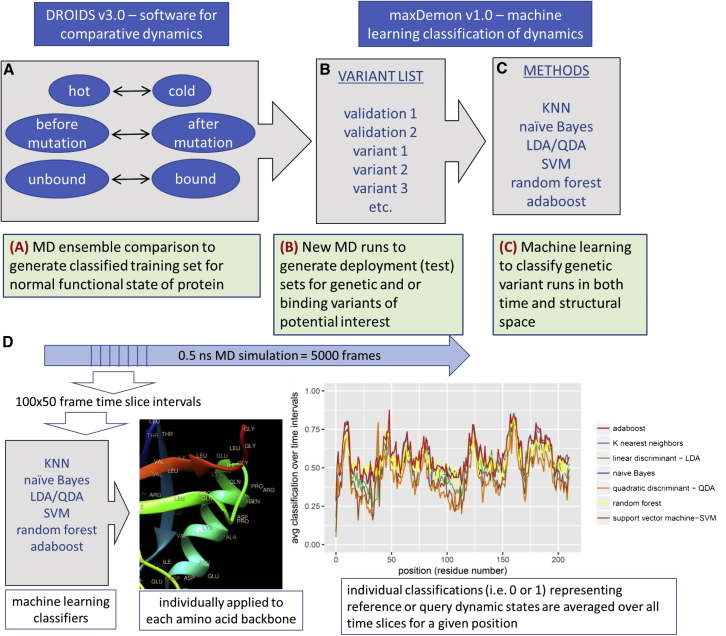
Figure 1.
Figure360 Schematic overview of DROIDS 3.0 + maxDemon 1.0 software for machine-learning-based detection of variant impacts on functionally conserved protein dynamics. The pipeline starts with (A) generation of two large ensembles of molecular dynamic (MD) simulations that represent a functional comparison of protein states (e.g., mutation, binding, or environmental change). The rmsf of protein backbone atoms in these ensembles are comparatively analyzed/visualized (i.e., using DROIDS) and are also later used as preclassified training data sets for machine learning (i.e., using maxDemon). Note: the pictured DROIDS analysis of nucleosome shows overall dampening of rmsf in the histone core with maximal dampening where the histone tails cross the DNA helix. (B) New MD simulations are generated on two structures self-similar to the query state of training as well as a list of functional variants, and (C) up to seven machine learning methods are employed to classify the MD in the self-similar and variant runs according to the functional comparison defined by the initial training step. (D) The performance of learning is defined by average value of classification (i.e., 0 or 1) over 50 frame time slices for each amino acid position, and regions of functionally conserved dynamics are later identified by significant canonical correlations in this learning efficiency (i.e., Wilk’s lambda) in self-similar MD validation runs. The impacts of variants are later defined by relative entropy of genetic/drug class variant MD compared with the MD in the self-similar runs (data not shown). To see this figure in color, go online.