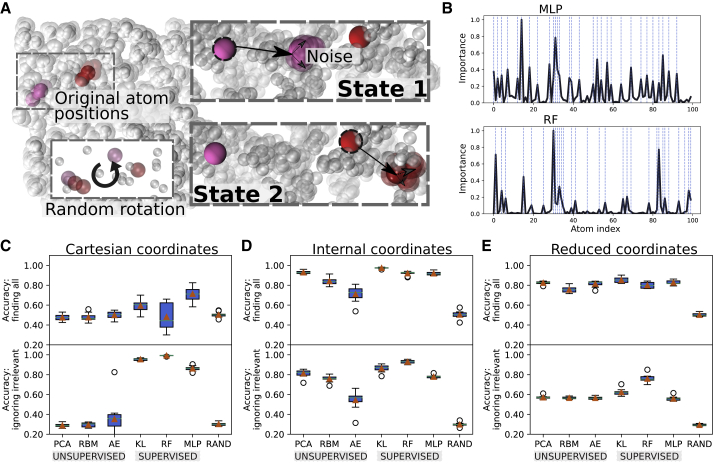
Figure 2.
(A) Toy model used to benchmark different ML approaches to extract important features from simulation data. Atomic coordinates are randomly generated, and a subset of the atoms, unique to every state, are displaced linearly from their initial positions. Artificial simulation frames are generated by adding noise to all atoms’ positions. Because only the relative and not the absolute positions of atoms are of significance in real biological systems, the system is rotated randomly around the origin. (B) Importance per atom for single instances of the toy model is shown. The index of the displaced atoms is highlighted as dashed vertical lines, coinciding with all peaks in importance in the case of an MLP and some of them for an RF. (C–E) Box plots of the performance of the different methods using either Cartesian coordinates (C), the full set of inverse interatomic distances (D), or a reduced set of inverse interatomic distances (E) are given as input features sampled over different instances of the toy model with linear displacement and 10% noise level. A high accuracy at finding all atoms signifies that every displaced atom has been identified as important and that other atoms have low importance. A high accuracy at ignoring irrelevant atoms signifies that only displaced atoms, although not necessarily all of them, have been marked important (Methods and Fig. S10). The best performing set of hyperparameters found after benchmarking every method (Figs. S4–S9) have been used. RAND stands for random guessing. The boxplots show the median (orange horizontal line), the interquartile range (box), the upper and lower whiskers (vertical lines), as well as the outliers (circles). To see this figure in color, go online.