Figure 5.
Recruitment of the NER Pathway in Response to DNA ADP-Ribosylation
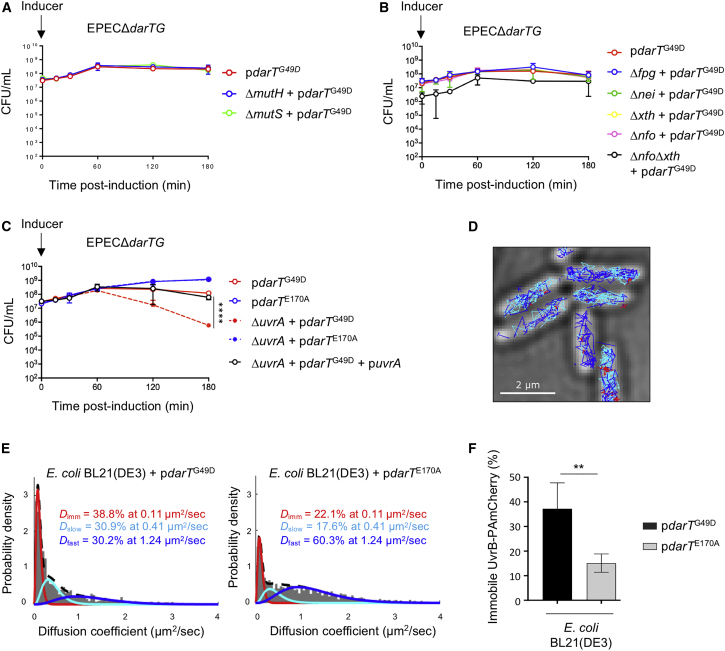
(A–C) Viability of EPECΔdarTG lacking single members of (A) the MMR pathway (EPECΔdarTGΔmutH and EPECΔdarTGΔmutS), (B) the BER pathway (EPECΔdarTGΔfpg and EPECΔdarTGΔnei, EPECΔdarTGΔxth, and EPECΔdarTGΔnfo), and (C) the NER pathway (EPECΔdarTGΔuvrA) following expression of darTG49D; n = 3 ± SD, ∗∗∗∗p < 0.00001 by two-way ANOVA with or without uvrA + pdarTG49D.
(D) Single-molecule tracking of UvrB-PAmCherry over 10,000 frames (with a 15 ms interval between frames) following expression of darTG49D. Immobile UvrB-PAmCherry molecules with D∗ < 0.2 μm2/s are in red, with 0.2 μm2/s < D∗ (the diffusion coefficient) < 1.5 μm2/s in turquoise and D∗ > 1.5 μm2/s in blue.
(E) Determination of D∗ values of UvrB-PAmCherry in E. coli, fitted with a three-species model following 15 min expression of darTG49D or darTE170A.
(F) Percentage of immobile UvrB-PAmCherry in E. coli after 15 min expression of darTG49D or darTE170A; n = 4 biological replicates ± SD, total number of cells = 14,338, total number of tracks = 142,748, ∗∗p < 0.01 by unpaired t test (two tailed).