Abstract
Homing based gene drives (HGD) possess the potential to spread linked cargo genes into natural populations and are poised to revolutionize population control of animals. Given that host encoded genes have been identified that are important for pathogen transmission, targeting these genes using guide RNAs as cargo genes linked to drives may provide a robust method to prevent disease transmission. However, effectiveness of the inclusion of additional guide RNAs that target separate genes has not been thoroughly explored. To test this approach, we generated a split-HGD in Drosophila melanogaster that encoded a drive linked effector consisting of a second gRNA engineered to target a separate host-encoded gene, which we term a gRNA-mediated effector (GME). This design enabled us to assess homing and knockout efficiencies of two target genes simultaneously, and also explore the timing and tissue specificity of Cas9 expression on cleavage/homing rates. We demonstrate that inclusion of a GME can result in high efficiency of disruption of both genes during super-Mendelian propagation of split-HGD. Furthermore, both genes were knocked out one generation earlier than expected indicating the robust somatic expression of Cas9 driven by Drosophila germline-limited promoters. We also assess the efficiency of ‘shadow drive’ generated by maternally deposited Cas9 protein and accumulation of drive-induced resistance alleles along multiple generations, and discuss design principles of HGD that could mitigate the accumulation of resistance alleles while incorporating a GME.
Keywords: CRISPR, Cas9, Homing, split-HGD, Drosophila melanogaster, resistance allele
For standard Mendelian inheritance, any particular allele has a 50% chance in being transmitted to its offspring. While mechanisms of meiosis generally bias selection against violators of Mendel’s rules, there are many examples of naturally occurring selfish genetic elements (SGEs) that succeed in bypassing these rules. These SGEs enhance, or “drive” their transmission into subsequent generations, despite often times being harmful to the harboring individual (i.e., imposing a fitness load). These include, for example, transposable elements (TEs), meiotic drivers, B chromosomes, post segregation killers, heritable microbes, and homing endonuclease genes (Werren et al. 1988; Burt and Trivers 2006; Werren 2011; McLaughlin and Malik 2017). Drawing inspiration from these natural systems, strategies for exploiting drive to alter the genetics of wild pest populations have been proposed (Werren et al. 1988; Burt, 2003; Burt and Trivers 2006; Werren 2011; Esvelt et al. 2014; Champer et al. 2016; McLaughlin and Malik 2017), and some have even been experimentally tested in the laboratory, however none have been implemented in the field. For those tested in the laboratory, some examples include synthetic Medea elements (Chen et al. 2007; Akbari et al. 2014; Buchman et al. 2018a), engineered underdominance systems (Akbari et al. 2013; Buchman et al. 2018b), and those whose development was accelerated by the CRISPR revolution (Jinek et al. 2012; Cong et al. 2013; Mali et al. 2013) including toxin-antidote based systems (Oberhofer et al. 2019), and homing based gene drive systems (HGDs) (Esvelt et al. 2014; Gantz and Bier 2016; Champer et al. 2016; Marshall and Akbari 2018).
HGDs are perhaps the furthest along in development, and have already been tested in a broad range range of organisms spanning bacteria, yeast, insects, and mammals (Windbichler et al. 2011; Gantz and Bier 2015; DiCarlo et al. 2015; Gantz et al. 2015; Hammond et al. 2016, 2018; Champer et al. 2017, 2018; KaramiNejadRanjbar et al. 2018; Kyrou et al. 2018; Yan and Finnigan 2018; Li et al. 2019; Grunwald et al., 2019, Valderrama et al., 2019). They function by encoding the Cas9 endonuclease and an independently expressed guide RNA (gRNA) responsible for mediating DNA/RNA base pairing and cleavage at a predetermined site (Esvelt et al. 2014; Gantz and Bier 2015, 2016; Champer et al. 2016; Marshall and Akbari 2018). When the HGD is positioned within its target site in a heterozygote, double stranded DNA breaks (DSBs) on the opposite chromosome can result in the drive allele being used as a template (i.e., donor chromosome) for DNA repair mediated by homologous recombination. This can result in copying, or “homing,” of the HGD into the broken chromosome (i.e., receiver chromosome), thereby converting heterozygotes to homozygotes in the germline, which can bias Mendelian inheritance ratios and result in an increase in HGD frequency in a population.
Given the recent progress toward developing HGDs in pest species such as mosquitoes (Gantz et al. 2015; Hammond et al. 2016, 2018; Kyrou et al. 2018; Li et al. 2019), there is significant enthusiasm regarding their potential use to control wild populations. For example, given the enormous burden mosquitoes pose on humans, the release of HGDs linked with effector genes inhibiting mosquito pathogen transmission (Isaacs et al. 2011; Jupatanakul et al. 2017; Buchman et al. 2019a; b) may lead to replacement of disease-susceptible mosquitoes with disease-resistant counterparts resulting in reduced pathogen transmission (i.e., population modification drive). Alternatively, HGDs targeting genes affecting the fitness of female mosquitoes could also spread, resulting in gradual population declines and potentially even elimination (i.e., population suppression drive) (Windbichler et al. 2008, 2011; Kyrou et al. 2018). Given these features, both modification and suppression drives possess the potential to transform mosquito population control measures (Burt 2003; Esvelt et al. 2014; Gantz and Bier 2016; Champer et al. 2016), and therefore have excited significant ongoing discussions involving their potential usage, regulation, safety, ethics and governance (Oye et al. 2014; Akbari et al. 2015; National Academies of Sciences, Engineering, and Medicine et al. 2016; Adelman et al. 2017). Despite these exciting developments however, the elephant in the room persists - can a gene drive actually work in the wild? There are a number of open questions looming as to the efficiency of HGDs. For example, can a drive spread to fixation in the wild? Will it simply breakdown due to resistance? Will the linked anti-pathogen effector work efficiently given the expected diversity of parasites/virus genomes found in the wild? Can the pathogen evolve to become resistant to the anti-pathogen effector and perhaps even become more virulent (Marshall et al. 2019)? These are just a minority of legitimate concerns regarding the potential use of a gene drive that would need to be resolved prior to any release.
While many questions loom, there has been some effort to resolve these concerns safely in the lab. For example, with regard to the HGD breakdown due to resistance, multiple studies have explored design criteria attempting to suppress the effects of resistance alleles on drive propagation. For example, some studies have had some success using germline-restricted promoters to express Cas9 increasing rates of HDR, resulting in increased homing rates, as opposed to error-prone pathways such as non-homologous end joining (NHEJ) which results in the generation of resistance alleles (Hammond et al. 2018; Champer et al. 2018). Other studies have described (Esvelt et al. 2014; Champer et al. 2016; Marshall et al. 2017) and tested (Champer et al. 2018, 2019a; b; Oberhofer et al. 2018) multiplexed gRNAs in drives resulting in moderate increases in drive efficacy. While others have had some success targeting highly conserved recessive fertility/viability genes whose homozygous mutants are inviable, or cannot reproduce, and therefore are expected to not affect the spread of HGDs (Hammond et al. 2016; KaramiNejadRanjbar et al. 2018; Kyrou et al. 2018; Oberhofer et al. 2018). However, despite these efforts, resistance alleles are still problematic, leaving open the question as to what is the best method to prevent their generation.
Here, to further explore this paramount issue of resistance to HGD we use Drosophila melanogaster as our model. We use a genetic safeguarded split-drive design as a safety feature and also encode a linked effector to the drive. This effector consisted of a second gRNA engineered to target a separate host encoded gene which we term a gRNA-mediated effector (GME) (Figure 1). Given that there are many host-encoded genes that are important for pathogen transmission (Cheng et al. 2016; Dong et al. 2018), one potential application of a HGD is to incorporate a cargo GME that targets a host encoded factor that is important for some aspect of pathogen transmission. If the GME is effective, then disruption of its target in the population should in principle occur as the drive spreads, thereby immunizing that population from pathogen transmission. Therefore, encoding a GME in a drive may be a useful feature going forward and worth further exploring. As a proof of concept to test the efficiency of a HGD linked GME, we designed both the drive and effector to target phenotypic genes which resulted in easily scorable recessive viable phenotypes. This novel drive architecture enabled us to test many germline Cas9 expressing promoters, while simultaneously measuring homing and cleavage efficiencies in both the germline and soma for both target genes over successive generations. While homing rates were modest, cleavage rates were high. For example, we determined that we can reproducibly achieve complete penetrance of somatic mosaic phenotypes for both target genes with up to 100% efficiency stemming from a combination of Cas9 maternal deposition and somatic expression. However, despite the robust cleavage efficiencies and impressive efficacy of the HGD linked GME, drive resistance alleles were still generated which would hinder spread. Given these results, alternative design principles are proposed that could potentially mitigate these issues while also incorporating a drive linked GME.
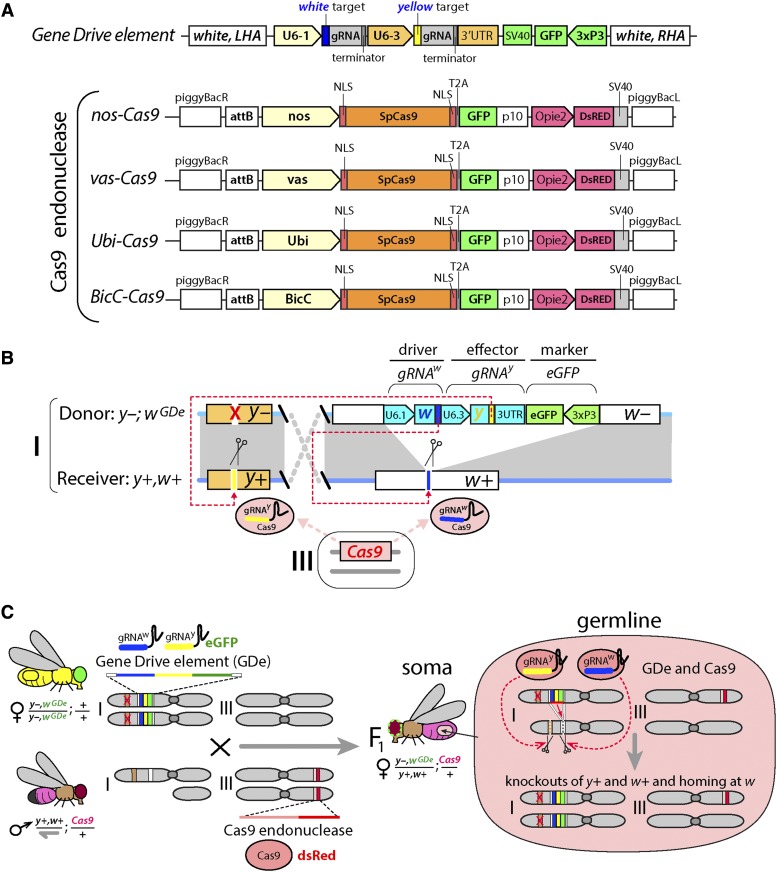
Figure 1.
Development of the CRISPR/Cas9-mediated split-drive system. The homing gene drive (HGD) system was split into two components: Gene Drive element (GDe) and Cas9 endonuclease (Cas9). (A) Schematic maps (not to scale) of genetic constructs used to assemble split-HGD systems. The GDe contains two guide RNAs (gRNAs) targeting the DNA cleavage at white and yellow loci, and an eye-specific marker (3xP3-GFP) all surrounded by Left and Right Homology Arms (LHA and RHA) complementary to the white cut site. Four Cas9 constructs expressing SpCas9 (Cas9) in early germline cells with nanos (nos) and vasa (vas) promoters, in late germ cells with Bicaudal C (BicC) promoter, and in both germ and somatic cells with Ubiquitin 63E (Ubi) promoter carried the eGFP linked to the coding sequence of Cas9 via a self-cleaving T2A sequence and a body specific marker of transgenesis (Opie2-DsRed). (B) GDe was site-specifically inserted at white locus on the 1st chromosome (i.e., X chromosome) in Drosophila via HDR-mediated integration, wGDe. The Cas9 constructs were inserted at the same site on the 3rd chromosome using φC31-mediated integration. In the presence of Cas9, GDe direct cleavage at both w+ and y+ loci and can home at white locus from the wGDe donor allele into the w+ receiver allele via HDR in heterozygotes. (C) The genetic cross between the GDe and Cas9 homozygous lines generates trans-heterozygous y–,wGDe/y+,w+; Cas9/+ females. The germline Cas9 expression is expected to limit the activity of the split-drive system, y+ and w+ knockouts and wGDe homing, to germ cells of the y–,wGDe/y+,w+; Cas9/+ females. The wGDe allele cannot home in Drosophila males, because they have only one X chromosome, aka. hemizygous.
Materials and Methods
Design and assembly of constructs
The genetic assembly of the Gene Drive element (GDe) with two gRNAs and 3xP3-eGFP (Figure 1A) was previously described to generate a split trans-complementing Gene Drive system (Lopez del Amo et al. 2019). The assembly of BicC-Cas9 construct followed the same steps previously described for the other three Cas9 lines: nos-Cas9, vas-Cas9, and Ubi-Cas9 (Kandul et al. 2019). The 2831 bases upstream of BicC-RA’s start codon (Bicaudal C, CG4824) was PCR amplified with CGACGGTCACGGCGGGCATGTCGACGCGGCCGCATAATTATATAATAATAAACTGCATGC (BicC-F) and TCCGTCGTGGTCCTTATAGTCCATGTTTAAACTGTGGAATTCGGATGATGATGATGATC (BicC-R) from Drosophila melanogaster genome, and enzymatically assembled (Gibson et al. 2009) into Ubi-Cas9 plasmid (addgene #112686) (Kandul et al. 2019) digested with NotI and XhoI.
Fly genetics and imaging
Flies were maintained under standard conditions at 25°. Embryo injections were carried at Rainbow Transgenic Flies, Inc. (http://www.rainbowgene.com). The BicC-Cas9 construct was inserted at the PBac{y+-attP-3B}KV00033 on the 3rd chromosome (Bloomington #9750) with φC31-mediated integration (Groth 2004). Transgenic flies were balanced with Df(3L)R/TM6C,cu1,Sb1,Tb1 (Bloomington #57) and CxD,ryBM/TM3,Sb1,Ser1 (Bloomington #1704) in the w+ genetic background.
To assess the cleavage rates and homing efficiencies of the split-drive system, we genetically crossed the GDe line to four different Cas9 lines in both directions. Two types of F1 trans-heterozygous y–,wGDe(eGFP)/y+,w+; Cas9(RFP)/+ females carrying either maternal or paternal Cas9 (F1 ♀ #2 or ♀ #4, respectively) and the F1 heterozygous y–,wGDe(eGFP)/y+,w+ females with either maternally or paternally deposited Cas9 protein were generated (F1 ♀ #1 or ♀ #3, respectively; Figure 2). Their yellow and white LOF mutations and transgene markers were scored. To explore whether yellow and white loci were also mutated in the germ cells of the F1 trans-heterozygous and heterozygous females, we genetically crossed them to w+,y+ and w–,y+ males, respectively, and examined their F2 progeny. LOF yellow mutations were scored only in male progeny that inherited their single X chromosome from mothers. To explore the behavior of resistance alleles over multiple generations, the F2 trans-heterozygous and heterozygous virgin female (♀ #6 or ♀ #5, respectively) progeny of F1 ♀ #2 were also collected, and genetic crosses and phenotype scoring were repeated for an additional generation, F3. The above crossing schemes are depicted in Figure 2. To generate means and standard deviations for statistical comparisons, each genetic cross was set up in triplicate using 10♂ and 10♀ flies for each replicate cross. Cleavage and homing frequencies are presented as percentages of y+ and w+ alleles in heterozygous females, aka. they normalized to 50% (Table S1).
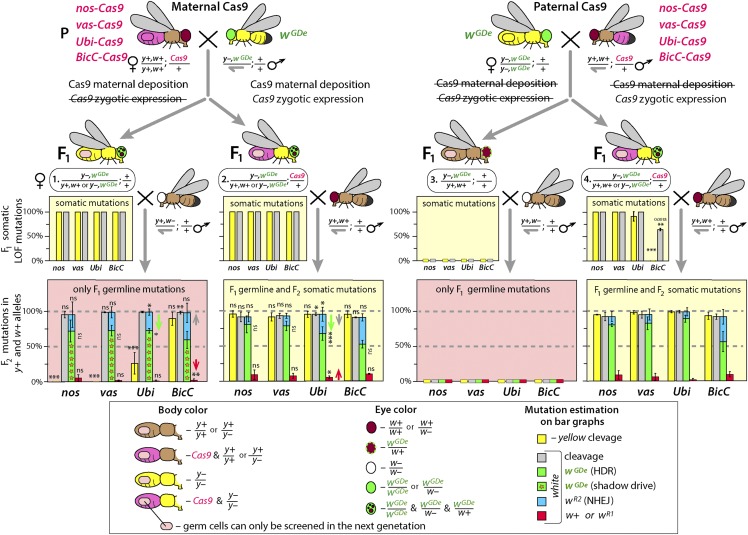
Figure 2.
Multiple Cas9 promoters induce double-gene knockouts in somatic tissues independently of maternal or paternal inheritance of Cas9. Each tested Cas9 promoter, including previously characterized germline-limited nos, vas, and BicC promoters, supported Cas9 expression in F1 somatic tissues and resulted in white and yellow loss-of-function (LOF) mutations in the F1 progeny. Furthermore, maternal deposition of Cas9 protein alone was sufficient to generate F1 somatic white and yellow LOF mutations as well as induce both homing (wGDe) and formation of resistance alleles (wR2) in w+ alleles of their germ cells (F1 ♀ #1). Paternal deposition of Cas9 protein did not induce mutations in somatic or germ cells (F1 ♀ #3). Notably, while 100% F1 parents had yellow LOF somatic mutations with each tested Cas9 line, only Ubi and BicC promoters deposited Cas9 protein sufficient to induce yellow LOF mutations in some germ cells (F1 ♀ #1). Zygotic expression of Cas9 under nos, vas, and Ubi promoters induced white and yellow LOF mutations in 100% trans-heterozygous females, while zygotic expression of BicC-Cas9 caused only white LOF mutation in 64.3% ± 2.6% of trans-heterozygous females (F1 ♀ #4). Rates of homing and resistance alleles were not significantly different among two types of trans-heterozygous (F1 ♀ #2 and #4) and heterozygous (♀ #1) females with maternally deposited Cas9. Only maternal deposition of Cas9 under Ubi promoter negatively affected homing rates (green arrows) in germ cells. Bar plots show the average ± SD over at least three biological replicate crosses. Statistical significance was estimated using a t-test with equal variance. (P ≥ 0.05ns, P < 0.05*, P < 0.01**, and P < 0.001***).
Flies were examined, scored, and imaged on the Leica M165FC fluorescent stereo microscope equipped with the Leica DMC2900 camera. To analyze Cas9 expression in ovaries of four homozygous Cas9 lines, their ovaries were dissected in PBS buffer, examined, and imaged utilizing the same settings. The eGFP fluorescence was used as a proxy of Cas9 expression, since it was tagged to Cas9 transgene via a T2A sequence (Figure S1).
Genotyping loci targeted with gRNAs
To explore the molecular changes that caused LOF and in-frame functional mutations in yellow and white loci, we PCR amplified the genomic regions containing target sites for gRNAw and gRNAy: GGCGATACTTGGATGCCCTGCGG and GGTTTTGGACACTGGAACCGTGG, respectively. Single-fly genomic DNA preps were prepared by homogenizing a fly in 30µl of a freshly prepared squishing buffer (10mM Tris-Cl pH 8.0, 1mM EDTA, 25mM NaCL, 200 μg/mL Proteinase K), incubating at 37° for 35 min, and heating at 95° for 2 min. 2 µl of genomic DNA was used as template in a 40 µL PCR reaction with LongAmp Taq DNA Polymerase (NEB). The 415bp PCR fragment of white target was amplified with CGTTAGGGAGCCGATAAAGAGGTCATCC (w.sF) and AAGAACGGTGAGTTTCTATTCGCAGTCGG (w.sR); and CACTCTGACCTATATAAACATGGACCGCAGTTTG (y.sF) and CCAATTCATCGGCAAAATAGGCATATGCAT (y.sR) primers were used to amplify the 375bp PCR fragment of yellow. PCR aplicons were purified using QIAquick PCR purification kit (QIAGEN), and sequenced in both directions with Sanger method at Source BioScience. To characterize molecular changes at the targeted sites, sequence AB1 files were aligned against the corresponding reference sequences in SnapGene 4.
Statistical analysis
Statistical analysis was performed in JMP 8.0.2 by SAS Institute Inc. At least three biological replicates were used to generate statistical means for comparisons. To estimate the effect of Cas9 maternal deposition on homing efficiency, rates of cleavage, homing, and resistance allele formation in F1 ♀ #4 with paternal Cas9 were compared to the corresponding values in F1 ♀ #1 and ♀ #2 with maternally deposited Cas9 protein (Figure 2). To assess the significance of resistance allele accumulation and homing rate decline between F2 and F3 generations, rates of cleavage, homing, and resistance alleles in F2 ♀ #5 and F2 ♀ #6 (Figure 3A) were compared to the corresponding values in F1 ♀ #1 and F1 ♀ #2, respectively (Figure 2). P values were calculated for a two-sample Student’s t-test with equal variance.
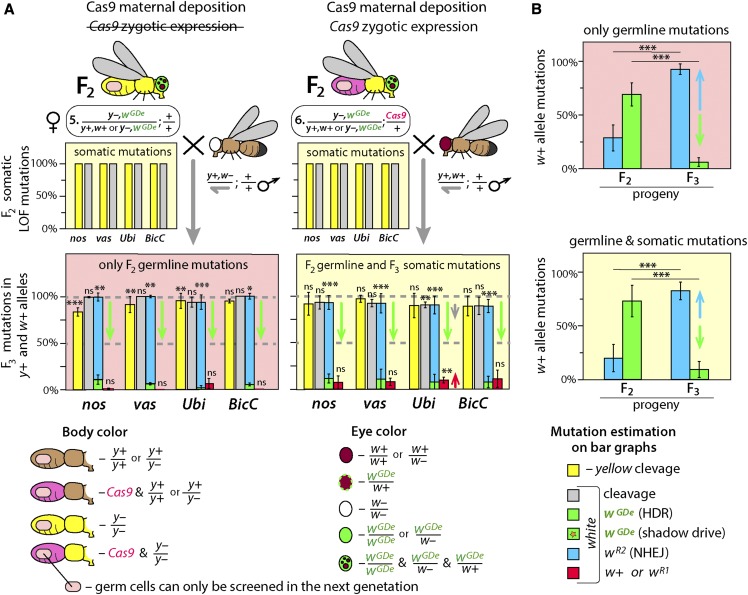
Figure 3.
Resistance alleles accumulates over subsequent generations and restricts homing. Resistance alleles are expected to be immune to the further cleavage by the same Cas9/gRNA system and if their carrier is fertile can propagate at the expense of homing. (A) To explore this phenomenon, F2 ♀ #5 and F2 ♀ #6 collected among progeny of F1 ♀ #2 were genetically crossed with w– and w+ males, respectively, and their F3 progeny were scored. While the cleavage rate in F2 germ cells decreased only in F2 ♀ #6 with Ubi-Cas9 (red arrow) likely due to the rise of functional wR1 alleles, the homing frequency fell significantly for each tested split-drive system with and without Cas9 gene (green arrows). The fall of homing rate was accompanied by the accumulation of the wR2 alleles. (B) Accumulation of wR2 alleles resistant to cleavage by Cas9/gRNAw restricted homing of GDe. Frequencies of homing and resistance alleles were averaged for all tested promoters and presented separately for progeny of heterozygous and trans-heterozygous females, F2 ♀ #5 and F2 ♀ #6, respectively. Resistance allele frequency increased from 28.5% or 19.9% to 92.6% or 82.6%, respectively, between F2 and F3 (blue arrows) and caused the dramatic decline in homing from 69.0% or 73.0% to 6.1% or 9.2%, respectively (green arrows). Notably, scoring of wR2 alleles in w– recessive background resulted in the higher estimation of white LOF mutations alleles, since wR2 alleles were complemented by w+ alleles inherited from wild type males. Bar plots show the average ± SD over at least three biological replicate crosses. Statistical significance was estimated using a t-test with equal variance. (P ≥ 0.05ns, P < 0.05*, P < 0.01**, and P < 0.001***).
Gene drive safety measures
All crosses using gene drives genetics were performed in accordance to an Institutional Biosafety Committee-approved protocol from UCSD in which full gene-drive experiments are performed in a high-security ACL2 barrier facility and split drive experiments are performed in an ACL1 insectary in plastic vials that are autoclaved prior to being discarded in accord with currently suggested guidelines for laboratory confinement of gene-drive systems (Akbari et al. 2015; National Academies of Sciences, Engineering, and Medicine et al. 2016).
Ethical conduct of research
We have complied with all relevant ethical regulations for animal testing and research and conformed to the UCSD institutionally approved biological use authorization protocol (BUA #R2401).
Data availability
All data that are represented fully within the tables and figures. The nos-, vas-, Ubi-Cas9 plasmids and the corresponding fly lines are deposited at Bloomington Drosophila Stock Center (#79004 – #79006) and AddGene.org (#112685 – #112687), respectively. The BicC-Cas9 and GDe plasmids and fly lines will be made available upon request. Supplemental material available at figshare: https://doi.org/10.25387/g3.11449542.
Results
Design of split-HGD encoding two gRNAs
To assess the feasibility and efficiency of utilizing a HGD to bias transmission while also expressing a GME, we designed a HGD that expressed two gRNAs (Lopez del Amo et al. 2019). The homing component of the split-HGD system, referred herein as a Gene Drive element (GDe), encodes a gRNA targeting white (gRNAw, driver gRNA), a separate cargo GME targeting yellow (gRNAy, effector), a 3xP3-eGFP dominant marker, all together flanked by 1kb homology arms from the white target locus to direct targeted HDR mediated integration (Figure 1A). The GDe was integrated at the white locus (wGDe) in D. melanogaster via HDR. In the presence of Cas9, the GDe directs cleavage at both white and yellow, both X-linked loci, and is also capable of homing into the white locus (Figure 1B-C). Importantly, in D. melanogaster homozygous loss-of-function (LOF) mutants of both white and yellow are viable and fertile with scorable recessive LOF phenotypes in the eye and body, respectively, enabling cleavage events to be directly quantified over successive generations. Additionally, males have only one X chromosome, and are therefore hemizygous for white and yellow, restricting the quantification of homing to heterozygous females (y–,wGDe/y+,w+, Figure 1C).
High penetrance of F1 somatic mutations generated by Cas9 through both maternal deposition and zygotic expression
We explored the effects of tissue specificity and timing of Cas9 expression on cleavage and homing in the germline by using four separate promoters with distinct expression profiles to express Cas9-T2A-GFP: nanos (nos) (Van Doren et al. 1998) and vasa (vas) promoters known for early germline-limited expression (Hay et al. 1988; Van Doren et al. 1998; Sano et al. 2002); Bicaudal C (BicC) promoter supporting later germline-limited expression (Saffman et al. 1998); and Ubiquitin 63E (Ubi) promoter with strong expression in both somatic and germ cells (Preston et al. 2006; Akbari et al. 2009). We controlled for variation in expression due to position effect (PE), by integrating each Cas9 construct (Figure 1A) into the same site on the 3rd chromosome using φC31-mediated integration (Groth 2004). We confirmed germline expression by imaging the expression of a self-cleaving T2A-eGFP tag attached to the coding sequence of Cas9, and each promoter robustly expressed GFP in the ovaries (Lowest nos-Cas9 < vas-Cas9 < Ubi-Cas9 < BicC-Cas9 Highest) (Figure S1).
We quantified cleavage efficiencies by performing bi-directional crosses between hemizygous or homozygous GDe lines mated to heterozygous Cas9 lines (Figure 2). From these crosses we determined that maternally deposited Cas9 protein is sufficient to induce both yellow and white somatic LOF mutations in F1 females heterozygous for the GDe both in presence (♀# 2; y–,wGDe/y+,w+; Cas9/+) and in the absence (♀ # 1; y–,wGDe/y+,w+; Figure 2) of Cas9 gene inheritance. To determine whether zygotic expression of Cas9 can also induce somatic mutations, we scored white and yellow LOF somatic mutations in F1 trans-heterozygous females inheriting Cas9 exclusively from their fathers (i.e., paternal Cas9). Unexpectedly, F1 trans-heterozygous female progeny inheriting Cas9 as a gene (♀#4; y–,wGDe/y+,w+; Cas9/+; Figure 2) from their fathers had mutations in both white and yellow with varying frequencies depending on which promoter drove Cas9 expression. For example, nos-Cas9 and vas-Cas9 – induced 100% white and yellow LOF somatic mutations in F1 trans-heterozygous females, while Ubi-Cas9 resulted in 100% of white and 91.3% ± 9.7% of yellow LOF somatic mutations, and BicC-Cas9 resulted in only white LOF mutations in 64.3% ± 2.6% of the F1 y–,wGDe/y+,w+; BicC-Cas9/+ progeny (Figure 2). Interestingly however, 100% of F1 heterozygous female progeny from the same fathers that did not inherit Cas9 as a gene (♀#3; y–,wGDe/y+,w+; +/+; Figure 2) had wild type (wt) phenotypes, for both white (red eyes) and yellow (brown body), presumably resulting from lack of sufficient Cas9 protein deposited paternally to induce mutations in the zygote (Table S1). Taken together these data indicate that the Cas9 promoters tested here are active both maternally and zygotically and can promote very high cleavage efficiency in somatic cells.
We assessed whether the yellow and white alleles were mutated by maternally deposited Cas9 in germ cells of F1 y–,wGDe/y+,w+ females by mating these females to y+,w– males and scored recessive yellow phenotypes in resulting F2 male progeny (y–) and recessive white phenotypes in resulting F2 male and female progeny (w–/w–). We found that maternally deposited Cas9 protein expressed under nos and vas promoters did not induce yellow LOF mutations in germ cells of F1 females, while expression from Ubi and BicC promoters resulted in 26% ± 15% and 89.4% ± 9.4% of yellow alleles being mutated in germ cells of F1 females (Figure 2), respectively, perhaps due to a stronger maternal deposition of Cas9 protein by these promoters (Figure S1) combined with possible preferential gRNA loading by Cas9. Despite the lack of LOF germline mutations in yellow by nos and vas, every tested Cas9 line provided a sufficient amount of maternally deposited Cas9 protein to knockout the white allele in 94.9% ± 4.5–98.8% ± 1.1% of F1 germ cells (measured in F2 progeny; Figure 2, Table S1). We explored whether the w+ alleles (1.2–5.1%) were cut by Cas9, and perhaps repaired into cleavage resistance alleles, by perfoming Sanger sequencing of PCR amplicons of the white target locus from individual male flies. Each tested F2 male with red eyes (w+) indeed had a wt w+ allele, and we did not find any white in-frame functional resistance alleles in F1 germ cells suggesting that these alleles likely remained uncut in the germline.
Maternally deposited Cas9 is sufficient to induce homing of GDe in germ cells
The Cas9/gRNAw-induced DSBs at white locus can be repaired either by HDR resulting in homing of the GDe (wGDe/wGDe) or NHEJ incorporating indel mutations that can render the target locus unrecognizable by the Cas9/gRNAw machinery, and when these mutations occur in germ cells they are referred to as resistance alleles (wR): here LOF and in-frame functional resistance alleles are referred as wR2 and wR1, respectively (Figure S2). To directly estimate the frequency of wGDe homing and wR generation in the absence of additional somatic mutations resulting from zygotic expression of Cas9, we analyzed white phenotypes in the F2 progeny of the F1 wGDe/w+ females with maternally deposited Cas9 in a w– recessive mutant background (Figure 2). Every tested Cas9 promoter provided a sufficient amount of maternally deposited Cas9 in the F1 germ cells to enable the conversion of 59–72% of w+ alleles into wGDe (i.e., homing of GDe) in y–,wGDe/y+,w+ females. This conversion which occurs in the presence of Cas9 protein, but absence of inheritance of the Cas9 gene, was previously noted and termed “shadow drive” (Guichard et al. 2019). The remaining DSBs at w+ alleles were repaired by NHEJ and generated around 38–23% wR2 alleles (Figure 2). To explore molecular changes at white locus, we PCR amplified and Sanger sequenced wR2 alleles from individual F2 male progeny and identified indels localized at the white cut site in each sequenced male (Figure S3A). The maternally deposited Cas9 by BicC promoter resulted in the lowest homing and the highest resistance allele rates (59.3% ± 12.3% and 38.7% ± 13.7%, respectively), though no significant difference was identified between BicC and other Cas9 promoters. Nevertheless, each tested promoter supplied Cas9 protein via mothers to the progeny that enabled shadow drive, thus resulting in super-Mendelian propagation of wGDe to their grandchildren.
Maternal deposition of Cas9 protein reduces the homing efficiency
Maternally deposited Cas9 can induce white cleavage and repair mediated by NHEJ as opposed to HDR in mitotically dividing germ cells which can result in a bias toward generating resistance alleles (wR2 and wR1) at the expense of homing wGDe (Lopez del Amo et al., 2019). To explore this effect, we compared homing rates between F1 trans-heterozygous females that inherited Cas9 either maternally (♀#2; y–,wGDe/y+,w+; Cas9/+) or paternally (♀#4; y–,wGDe/y+,w+; Cas9/+; Figure 2). For nos-Cas9, vas-Cas9, and BicC-Cas9, maternal deposition of Cas9 did not result in a significant bias in homing efficiencies. However, for Ubi-Cas9 homing rates were significantly lower (67%) in the trans-heterozygous females that inherited Cas9 maternally (♀#2; wGDe/w+; Ubi-Cas9/+) as compared to 88% for trans-heterozygous females inheriting Ubi-Cas9 paternally (♀#4; wGDe/w+; Ubi-Cas9/+). In addition to the lower homing rates for Ubi-Cas9, the rate of wR2 alleles was significantly higher with maternally deposited Ubi-Cas9 as compared to paternally deposited Ubi-Cas9: 9.9% ± 5.7% vs. 27.3% ± 10.0%, P > 0.025 or 26.5% ± 4.4%, P > 0.029, respectively (Figure 2). Taken together, these results suggest that high levels of maternal deposition of Cas9 protein into developing oocytes can result in white cleavage in mitotic cells, prior to developmental stages where efficient HDR repair occurs, therefore leading to a higher frequency of wR events.
Resistance alleles accumulate between F2 and F3 generations
Resistance alleles generated in germ cells are immune to subsequent cleavage by the Cas9/gRNAw complex. Drosophila white and yellow LOF homozygotes are viable and fertile, as a result, the frequency of resistance alleles can potentially increase from generation to generation. We explored this possibility by crossing F2 trans-heterozygous females (♀ #6, wGDe/w+; Ubi-Cas9/+; Figure 3A) to wt (y+,w+) males, and scored their F3 progeny for yellow and white phenotypes, as well as for inheritance of the GDe. Indeed, the frequency of white LOF mutations (wR2) increased significantly between F2 and F3 progenies for each Cas9 promoter: 11.2% ± 6.2% vs. 81.7% ± 7.5% for nos-Cas9; 13.2% ± 5.6% vs. 82.4% ± 10.4% for vas-Cas9; 18.6% ± 12.0% vs. 84.6% ± 9.5% for Ubi-Cas9; and 36.7% ± 7.5% vs. 81.6% ± 7.1% for BicC-Cas9, P > 0.0001, respectively. This increased frequency of generating wR2 alleles negatively affected the homing rate, which dropped between F2 and F3 generations: from 80.0% ± 7.7% to 11.3% ± 4.8% for nos-Cas9; from 80.2% ± 7.4% to 10.8% ± 10.4% for vas-Cas9; from 78.0% ± 13.2% to 7.4% ± 8.4% for Ubi-Cas9; and from 53.9% ± 9.8% to 7.6% ± 6.2% for BicC-Cas9 (♀ #6, wGDe/w+; Ubi-Cas9/+; Figure 3A). To avoid any ambiguity caused by somatic expression of Cas9, the same analysis was repeated with the F2 heterozygous females carrying maternally deposited Cas9 protein but lacking the Cas9 gene resulting in similar conclusions (♀ #5, y–,wGDe/y+,w+; Figure 3A). We assessed the accumulation of resistance alleles by comparing the mean frequencies of homing and resistance alleles between F2 and F3 generations. The frequency of resistance alleles rose from 28.5% ± 12.2% to 92.6% ± 5.0% in heterozygous females or from 19.9% ± 12.8% to 82.6% ± 8.2% in trans-heterozygous females, and decreased the homing rate from 69.0% ± 10.8% to 6.1% ± 4.2% and from 73.0% ± 14.6% to 9.2% ± 7.5%, respectively (P > 0.0001, Figure 3B). As expected, the frequency of LOF resistance alleles at white locus (wR1) also increased from F2 to F3 generations and further restricted homing of the GDe. The frequency of in-frame functional white and yellow mutations (wR1 and yR1) could also increase in the F3 progeny, but unfortunately this effect could not be directly estimated. The frequency of cleavage at white significantly decreased in the F3 progeny of F2 y–,wGDe/y+,w+; Ubi-Cas9/+ females, and could be explained by the increase of wR1 allele rate that were indistinguishable from w+ alleles phenotypically: from 3.4% ± 2.6% in F2 to 9.7% ± 3.1% in F3, P > 0.004 (Figures 2, 3A). We tested this hypothesis by Sanger sequencing F3 wt males with red eyes and brown bodies, and identified in-frame indels and substitutions in the majority of tested males for each Cas9 promoter (wR1 and yR1 alleles, Figure S3). Therefore, many germ cells of F2 trans-heterozygous and heterozygous with maternally deposited Cas9 females had indel mutations in the white and yellow loci (y–,wGDe/yR,wR) that were indeed resistant to further cleavage by Cas9/gRNAw and Cas9/gRNAy, respectively.
Discussion
Homing based gene drives require efficient cleavage and copying in the germline in order to bias their transmission and are therefore sensitive to both existing and induced target sequence variation. In fact, the NHEJ-mediated generation of resistance alleles in germ cells was previously identified as the major force opposing the spread of HGD into populations (Gantz et al. 2015; Champer et al. 2017; Hammond et al. 2017; Oberhofer et al. 2018). Here, we used a split-drive design to further explore the effect of timing and location of Cas9 expression on both homing and resistance allele formation. This experimental design enabled us to separate effects of somatic expression from maternal deposition of Cas9 on the GDe inheritance and mutagenesis of a targeted gene. Additionally, we linked a GME to the GDe to measure the efficacy of the knockout of an additional gene, yellow. Using this approach, we were able to draw several conclusions, including; i) in addition to germline expression, each tested Cas9 promoter (nos, vas, BicC, Ubi) directs significant expression in somatic tissues; ii) the maternal protein deposition or gene expression of Cas9 is sufficient for homing (shadow drive) in germ cells; iii) paternal Cas9 protein deposition in the sperm is insufficient for the mutagenesis of a target gene; iv) drive-induced resistance alleles accumulate over generations and are predicted to restrict the spread of the drive; and v) expression of a drive mediating gRNA in addition to a linked GME can result in 100% penetrance of both scorable LOF phenotypes. Below we discuss these conclusions further and also propose novel drive architectures to potentially overcome these issues.
Somatic expression of Cas9 results in high mutagenesis rates
The maternal protein deposition and gene expression of Cas9 in the presence of a gRNA transgene were previously reported to induce LOF mutations in some F1 progeny from a cross using nos- or vas-driven Cas9 and U6-gRNA lines (Port et al. 2014; Lin and Potter 2016; Oberhofer et al. 2018; Kandul et al. 2019); however, the somatic nature of F1 LOF mutations was not fully explored. This is in part due to the fact that when Cas9 and gRNA are linked together in a single-locus HGD, somatic and germline LOF mutations are not easily distinguishable from heritable mutations occurring in prior generations, which can result in overestimation of mutation rates. Therefore, unlinking these components enables a better method for methodically disentangling these events. Here, using a split-drive design, we were able to carefully assess the effects of timing, expression, and inheritance of Cas9 on both homing and cleavage efficiencies. As reported previously, we found that maternal Cas9 protein deposition was sufficient to induce homing in germ cells, aka. shadow drive (Champer et al. 2019c; Guichard et al. 2019), in addition to high rates of F1 somatic LOF mutations (Port et al. 2014; Lin and Potter 2016; Oberhofer et al. 2018; Kandul et al. 2019). Interestingly, our estimations of homing rates by the shadow drive in white locus are notably higher than the previously reported in yellow locus: 59–72% (Figure 2) vs. 29–32% (Guichard et al. 2019) and 38% (Champer et al. 2019c), although the ratio of first generation drive to shadow drive frequencies is comparable in these systems (∼50%). The differences may be due to the yellow locus being less accessible to cleavage and HDR than white locus, or perhaps the lower fitness of yellow LOF somatic mutations (see below) biases against their inheritance (Massey et al. 2019).
Rather unexpectedly, we found that the zygotic expression of Cas9 alone (paternal Cas9), without the maternally deposited Cas9 protein, was also sufficient to induce F1 LOF somatic mutations. In fact, both nanos and vasa promoters, which were previously characterized to have early germline-limited expression (Van Doren et al. 1998; Sano et al. 2002; Kondo and Ueda 2013), in our system do support significant somatic expression of Cas9 which may stem from PE or perhaps the use of the P10 3′UTR. For example, F1 progeny with both maternally deposited Cas9 protein or with zygotically expressed Cas9 gene inherited from their fathers had white and yellow LOF somatic mutations with up to 100% efficiency (Figure 2). Consistent observations were reported in a recent work using a trans-complementing Gene Drive (tGD) system (Lopez del Amo et al. 2019) and for somatically induced lethality of Notch alleles driven by paternally provided Cas9 (Guichard et al., 2019). Taken together, these data conclusively demonstrate that, in the context of the tested promoters, Cas9 somatic expression confounds the estimation of mutagenesis rates in germ cells and can result in the overestimation of homing rates for a single-locus HGD.
Resistance alleles accumulate over subsequent generations
Consistent with previous studies, we found that maternal deposition of Cas9 protein into embryos inheriting a GDe results in both resistance allele formation and homing in the germ cells (Champer et al. 2019c; Guichard et al. 2019). In addition to this observation, we also found that paternal Cas9 protein deposition was not sufficient to induce mutagenesis in target genes, presumably due to the low quantities of Cas9 carried by the sperm into the egg. Moreover, we determined that maternal deposition of Cas9 protein in trans-heterozygous females with vas- and nos-Cas9 does not induce more resistance alleles at the expense of a homing rate than those in the females that inherit vas- and nos-Cas9 paternally.
In the light of substantial somatic expression of Cas9 driven by common Drosophila germline promoters (Figure 2), the germline inheritance rate provides a better estimate of the rate of inducing resistance alleles in germ cells than the ‘embryonic resistance allele’ frequency used previously (Champer et al. 2019c). Consequently, our estimations of F2 resistance allele formation are lower than those reported by Champer et al. as the embryo R2 (LOF) resistance alleles for a split-gene drive system with the nos-Cas9, 11% ± 6% (Figure 2) vs. 74% ± 2% (Champer et al. 2019c).
The frequency of resistance alleles (wR) increased dramatically between F2 and F3 generations and correlated with decreases in homing (Figure 3). Taken together, these results suggest that HDR-mediated homing and NHEJ-mediated formation of resistance alleles are integral outcomes of DSBs repair induced by Cas9/gRNA; and when resistance alleles do not cause lethality or sterility to their carrier, the accumulation of resistance alleles is predicted to impede the spread of the drive (Hammond et al. 2017; KaramiNejadRanjbar et al. 2018; Oberhofer et al. 2018).
gRNA-mediated effector (GME)
The CRISPR/Cas9 technology was previously used in the combination with multiple gRNAs to knock out or convert different genes simultaneously (e.g., Cong et al. 2013; Lopez del Amo et al. 2019; Kandul et al. 2019; Guichard et al. 2019). Here we linked the second gRNA targeting yellow to the GDe inside the white locus, and demonstrated that both yellow and white were effectively knocked out in heterozygous females. In principle, the GME approach can be used to knock out multiple genes located on different chromosomes, such as multiple host factors required for mosquito infection with pathogens or repressors of mosquito anti-pathogen immune genes (Simões et al. 2018). Unlike the the allelic drive (Guichard et al. 2019), the GME does not require the HDR-mediated conversion in germ cells; instead, it relies on the NHEJ-mediated indel formation in somatic tissues, and the widespread Cas9 somatic expression described here is expected to improve the penetrance of the GME-mediated knockout (Kandul et al. 2019). Robust knockout of host genes in somatic tissues may reduce the mosquito fitness (Dong et al. 2018), but an efficient gene drive can spread its cargo genes in a population even if they are costly to their carriers (Kyrou et al. 2018). Therefore, our results suggest that the GME directing knock out of multiple mosquito genes to suppress pathogen infection in mosquitoes may be a viable strategy and should be further explored going forward.
Novel strategies for disarming resistance alleles in germ cells
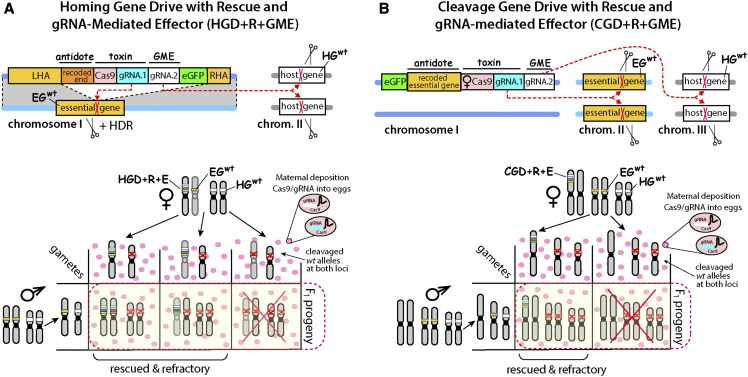
The accumulation of drive resistance alleles reported here was in part due to the fact that white is recessive viable, enabling NHEJ-induced resistance alleles to accumulate. Given this accumulation, targeting non-essential genes using HGD may not be ideal. To avoid this issue, targeting essential genes would be a more appropriate design to ensure gene drive stability and spread. By targeting essential genes, it is possible that non-drive resistance alleles could be actively selected against using a phenomenon previously termed as lethal mosaicism (Kandul et al. 2019; Guichard et al. 2019) or by natural selection due to increased fitness costs. Lethal mosaicism results in dominant biallelic knockouts of target genes throughout development, which could eliminate cleavage resistance alleles as they would be non-viable. We envision two novel drive design architectures that incorporate a GME and rely on lethal mosaicism to limit the generation of resistance alleles. First, haplo-sufficient genes essential for insect viability or fertility can be targeted by HGD designed to express a recoded version of the disrupted gene that is resistant to gRNA-mediated cleavage in addition to a linked GME (HGD+R+GME). This ensures that only the progeny that inherit the HGD+R+GME survive, while all progeny that inherit a cleaved allele perish due to non-rescued lethal mosaicism (Figure 4). Second, a Cleavage-only Gene Drive with Rescue could be designed that incorporates a GME (CGD+R+GME) which mechanistically relies exclusively on cleavage for biased inheritance and selection against drive resistance alleles (Figure 4)(Oberhofer et al. 2019). Both of these strategies would likely be effective in limiting the accumulation of drive resistance alleles. However, in-frame functional mutations (R1 type) that confer resistance against the Cas9/gRNA and do not cause fitness costs to carriers may still be generated, which could still limit the spread of a drive, and generation of these R1 alleles could possibly be further minimized by inclusion of additional gRNAs that target the essential genes to mediate drive. To summarize, our results demonstrate that inserting a GME into a HGD, efficient knockouts of multiple genes can be achieved while simultaneously biasing GDe transmission rates into subsequent generations. However, resistance alleles were generated, and accumulated, which would limit the efficacy and spread of this system. To overcome these limitations, novel drive architectures are proposed and remain to be tested in future studies.
Figure 4.
gRNA-mediated effector (GME) incorporated into two novel gene drive designs mechanistically based on lethal biallelic mosaicism. (A) Schematic of Homing Gene Drive targeting an essential gene with a recoded Rescue and GME (HGD+R+GME). The HGD+R+GME expresses Cas9 and two gRNAs targeting an essential gene (EG) and host gene (HG), a marker gene (eGFP), and the cleavage-resistant recorded portion of the essential gene that is being targeted by the gRNA/Cas9 complex (Rescue), which can rescue the knockout phenotype, flanked by Left and Right Homology Arms (LHA and RHA). Mechanistically, once HGD+R+GME is integrated precisely inside the EG it will direct cleavage of the EGwt allele on a receiver chromosome, and induce knockout mutations that will either result in lethal biallelic mosaicism, or convert the receiver chromosome into EGHGD+R+GME via homology directed repair (HDR). This ensures that only the progeny that inherit EGHGD+R+GME survive, while all progeny that inherit a cleaved EG allele perish due to non-rescued lethal mosaicism. In addition, the HGD+R+GME induces knockout of HG located on another (or the same) chromosome, leading to desired phenotype (i.e., pathogen resistance) to its carriers. The Punnett square below depicts the genetics of how HGD+R+GME achieves a 100% transmission rate and refractoriness in F1 progeny. Female heterozygous for HGD+R+GME maternally deposits Cas9/gRNA complexes into every oocyte knocking out both EG and HG, and only zygotes that inherit the HDR+R+GME would survive as F1 progeny. Notably, HDR will convert EGwt alleles into EGHGD+R+GME alleles and further increase numbers of surviving F1 progeny and this non-Mendelian inheritance rate will depend on homing efficiencies. (B) Schematic of Cleavage-only Gene Drive targeting an essential gene with a recoded Rescue and GME (CGD+R+GME). The CGD+R+GME expresses Cas9 with multiple gRNAs targeting an ES (gRNA.1) and HG (gRNA.2), a marker gene (eGFP), and the cleavage-resistant recorded essential gene (Rescue) integrated at a separate genomic location from the target gene. Mechanistically, a CGD+R+GME drive relies exclusively on cleavage with no HDR required for biased inheritance. A Punnett square depicts the genetics of how CGD+R+GME achieves 100% transmission and infection resistance rates in F1 progeny. The female heterozygous for CGD+R+GME deposits Cas9/gRNA complexes into every oocyte, only the half of the zygotes that inherits the CDR+R+MGE in a Mendelian fashion survive as F1 progeny, while the other half that do not inherit CDR+R+GME perishes due to lethal biallelic mosaicism.
Acknowledgments
This work was supported in part by the University of California, San Diego, Department of Biological Sciences, by funding from the Defense Advanced Research Project Agency (DARPA) under a “Safe Genes” Program Grant (HR0011-17-2-0047) awarded to O.S.A. and by the Office of the Director of the National Institutes of Health under award number DP5OD023098 awarded to V.M.G.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.11449542.
Communicating editor: R. Kulathinal
Literature Cited
- Adelman Z., Akbari O., Bauer J., Bier E., Bloss C. et al. , 2017. Rules of the road for insect gene drive research and testing. Nat. Biotechnol. 35: 716–718. 10.1038/nbt.3926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari O. S., Oliver D., Eyer K., and Pai C.-Y., 2009. An Entry/Gateway cloning system for general expression of genes with molecular tags in Drosophila melanogaster. BMC Cell Biol. 10: 8 10.1186/1471-2121-10-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari O. S., Matzen K. D., Marshall J. M., Huang H., Ward C. M. et al. , 2013. A Synthetic Gene Drive System for Local, Reversible Modification and Suppression of Insect Populations. Curr. Biol. 23: 671–677. 10.1016/j.cub.2013.02.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari O. S., Chen C.-H., Marshall J. M., Huang H., Antoshechkin I. et al. , 2014. Novel synthetic Medea selfish genetic elements drive population replacement in Drosophila; a theoretical exploration of Medea-dependent population suppression. ACS Synth. Biol. 3: 915–928. 10.1021/sb300079h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari O. S., Bellen H. J., Bier E., Bullock S. L., Burt A. et al. , 2015. BIOSAFETY. Safeguarding gene drive experiments in the laboratory. Science 349: 927–929. 10.1126/science.aac7932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A., Marshall J. M., Ostrovski D., Yang T., and Akbari O. S., 2018a Synthetically engineered Medea gene drive system in the worldwide crop pest Drosophila suzukii. Proc. Natl. Acad. Sci. USA 115: 4725–4730. 10.1073/pnas.1713139115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. B., Ivy T., Marshall J. M., Akbari O. S., and Hay B. A., 2018b Engineered Reciprocal Chromosome Translocations Drive High Threshold, Reversible Population Replacement in Drosophila. ACS Synth. Biol. 7: 1359–1370. 10.1021/acssynbio.7b00451 [DOI] [PubMed] [Google Scholar]
- Buchman A., Gamez S., Li M., Antoshechkin I., Lee S.-H. et al. , 2019a Broad Dengue Neutralization in Mosquitoes Expressing an Engineered Antibody, BioRxiv. 10.1101/645481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A., Gamez S., Li M., Antoshechkin I., Li H.-H. et al. , 2019b Engineered resistance to Zika virus in transgenic Aedes aegypti expressing a polycistronic cluster of synthetic small RNAs. Proc. Natl. Acad. Sci. USA 116: 3656–3661. 10.1073/pnas.1810771116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A., 2003. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. Biol. Sci. 270: 921–928. 10.1098/rspb.2002.2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A., and Trivers R., 2006. Genes in conflict: the biology of selfish genetic elements. Harvard University Press. 605p. 10.4159/9780674029118 [DOI]
- Champer J., Buchman A., and Akbari O. S., 2016. Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nat. Rev. Genet. 17: 146–159. 10.1038/nrg.2015.34 [DOI] [PubMed] [Google Scholar]
- Champer J., Reeves R., Oh S. Y., Liu C., Liu J. et al. , 2017. Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations. PLoS Genet. 13: e1006796 10.1371/journal.pgen.1006796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champer J., Liu J., Oh S. Y., Reeves R., Luthra A. et al. , 2018. Reducing resistance allele formation in CRISPR gene drive. Proc. Natl. Acad. Sci. USA 115: 5522–5527. 10.1073/pnas.1720354115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champer J., Yang E., Lee Y. L., Liu J., Clark A. G., et al. , 2019a Resistance is futile: A CRISPR homing gene drive targeting a haplolethal gene. BioRxiv. 10.1101/651737 [DOI] [PMC free article] [PubMed]
- Champer S. E., Oh S. Y., Liu C., Wen Z., Clark A. G., et al. , 2019b Computational and experimental performance of CRISPR homing gene drive strategies with multiplexed gRNAs. BioRxiv. 10.1101/679902 [DOI] [PMC free article] [PubMed]
- Champer J., Chung J., Lee Y. L., Liu C., Yang E. et al. , 2019c Molecular safeguarding of CRISPR gene drive experiments. eLife 8: e41439. 10.7554/eLife.41439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-H., Huang H., Ward C. M., Su J. T., Schaeffer L. V. et al. , 2007. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science 316: 597–600. 10.1126/science.1138595 [DOI] [PubMed] [Google Scholar]
- Cheng G., Liu Y., Wang P., and Xiao X., 2016. Mosquito Defense Strategies against Viral Infection. Trends Parasitol. 32: 177–186. 10.1016/j.pt.2015.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R. et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo J. E., Chavez A., Dietz S. L., Esvelt K. M., and Church G. M., 2015. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat. Biotechnol. 33: 1250–1255. 10.1038/nbt.3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Simões M. L., Marois E., and Dimopoulos G., 2018. CRISPR/Cas9 -mediated gene knockout of Anopheles gambiae FREP1 suppresses malaria parasite infection. PLoS Pathog. 14: e1006898 10.1371/journal.ppat.1006898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt K. M., Smidler A. L., Catteruccia F., and Church G. M., 2014. Concerning RNA-guided gene drives for the alteration of wild populations. eLife 3: e03401. 10.7554/eLife.03401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz V. M., and Bier E., 2015. Genome editing. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science 348: 442–444. 10.1126/science.aaa5945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz V. M., Jasinskiene N., Tatarenkova O., Fazekas A., Macias V. M. et al. , 2015. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. USA 112: E6736–E6743. 10.1073/pnas.1521077112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz V. M., and Bier E., 2016. The dawn of active genetics. BioEssays 38: 50–63. 10.1002/bies.201500102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A. 3rd et al. , 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Groth A. C., 2004. Construction of Transgenic Drosophila by Using the Site-Specific Integrase From Phage C31. Genetics 166: 1775–1782. 10.1534/genetics.166.4.1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald H. A., Gantz V. M., Poplawski G., Xu X.-R. S., Bier E. et al. , 2019. Super-Mendelian inheritance mediated by CRISPR–Cas9 in the female mouse germline. Nature 566: 105–109. 10.1038/s41586-019-0875-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard A., Haque T., Bobik M., Xu X.-R. S., Klanseck C. et al. , 2019. Efficient allelic-drive in Drosophila. Nat. Commun. 10: 1640 10.1038/s41467-019-09694-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond A., Galizi R., Kyrou K., Simoni A., Siniscalchi C. et al. , 2016. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 34: 78–83. 10.1038/nbt.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond A. M., Kyrou K., Bruttini M., North A., Galizi R. et al. , 2017. The creation and selection of mutations resistant to a gene drive over multiple generations in the malaria mosquito. PLoS Genet. 13: e1007039 10.1371/journal.pgen.1007039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond A. M., Kyrou K., Gribble M., Karlsson X., Morianou I., et al. , 2018. Improved CRISPR-based suppression gene drives mitigate resistance and impose a large reproductive load on laboratory-contained mosquito populations. BioRxiv. 10.1101/360339 [DOI]
- Hay B., Jan L. Y., and Jan Y. N., 1988. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell 55: 577–587. 10.1016/0092-8674(88)90216-4 [DOI] [PubMed] [Google Scholar]
- Isaacs A. T., Li F., Jasinskiene N., Chen X., Nirmala X. et al. , 2011. Engineered resistance to Plasmodium falciparum development in transgenic Anopheles stephensi. PLoS Pathog. 7: e1002017 10.1371/journal.ppat.1002017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A. et al. , 2012. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337: 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupatanakul N., Sim S., Angleró-Rodríguez Y. I., Souza-Neto J., Das S. et al. , 2017. Engineered Aedes aegypti JAK/STAT Pathway-Mediated Immunity to Dengue Virus. PLoS Negl. Trop. Dis. 11: e0005187 10.1371/journal.pntd.0005187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandul N. P., Liu J., Sanchez C H. M., Wu S. L., Marshall J. M. et al. , 2019. Transforming insect population control with precision guided sterile males with demonstration in flies. Nat. Commun. 10: 84 10.1038/s41467-018-07964-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- KaramiNejadRanjbar M., Eckermann K., Ahmed H. M. M., Sánchez C H. M., Dippel S. et al. , 2018. Consequences of resistance evolution in a Cas9-based sex conversion-suppression gene drive for insect pest management. Proc. Natl. Acad. Sci. USA 115: 6189–6194. 10.1073/pnas.1713825115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S., and Ueda R., 2013. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195: 715–721. 10.1534/genetics.113.156737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrou K., Hammond A. M., Galizi R., Kranjc N., Burt A. et al. , 2018. A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 36: 1062–1066. 10.1038/nbt.4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M., T. Yang, N. P. Kandul, M. Bui, S. Gamez, et al., 2019 Development of a Confinable Gene-Drive System in the Human Disease Vector, Aedes aegypti.BioRxiv. 10.1101/645440 [DOI] [PMC free article] [PubMed]
- Lin C.-C., and Potter C. J., 2016. Non-Mendelian Dominant Maternal Effects Caused by CRISPR/Cas9 Transgenic Components in Drosophila melanogaster. G3 (Bethesda) 6: 3685–3691. 10.1534/g3.116.034884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez del Amo, V. A. L. Bishop, H. M. Sanchez C., J. B. Bennett, et al, 2019 Split-gene drive system provides flexible application for safe laboratory investigation and potential field deployment. BioRxiv. 10.1101/684597 [DOI] [PMC free article] [PubMed]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M. et al. , 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J. M., Buchman A., Sánchez C H. M., and Akbari O. S., 2017. Overcoming evolved resistance to population-suppressing homing-based gene drives. Sci. Rep. 7: 3776 10.1038/s41598-017-02744-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J. M., and Akbari O. S., 2018. Can CRISPR-Based Gene Drive Be Confined in the Wild? A Question for Molecular and Population Biology. ACS Chem. Biol. 13: 424–430. 10.1021/acschembio.7b00923 [DOI] [PubMed] [Google Scholar]
- Marshall J. M., Raban R., Kandul N. P., Edula J. R., León T. et al. , 2019. Winning the tug-of-war between effector gene design and pathogen evolution in vector population replacement strategies. Front. Genet. 10: 1072 10.3389/fgene.2019.01072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey J. H., Chung D., Siwanowicz I., Stern D. L., and Wittkopp P. J., 2019. The yellow gene influences Drosophila male mating success through sex comb melanization. eLife 8: e49388. 10.7554/eLife.49388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin R. N. Jr, and Malik H. S., 2017. Genetic conflicts: the usual suspects and beyond. J. Exp. Biol. 220: 6–17. 10.1242/jeb.148148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Academies of Sciences, Engineering, and Medicine, Division on Earth and Life Studies, Board on Life Sciences, and Committee on Gene Drive Research in Non-Human Organisms: Recommendations for Responsible Conduct, 2016 Gene Drives on the Horizon: Advancing Science, Navigating Uncertainty, and Aligning Research with Public Values. National Academies Press. [PubMed]
- Oberhofer G., Ivy T., and Hay B. A., 2018. Behavior of homing endonuclease gene drives targeting genes required for viability or female fertility with multiplexed guide RNAs. Proc. Natl. Acad. Sci. USA 115: E9343–E9352. 10.1073/pnas.1805278115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhofer G., Ivy T., and Hay B. A., 2019. Cleave and Rescue, a novel selfish genetic element and general strategy for gene drive. Proc. Natl. Acad. Sci. USA 116: 6250–6259. 10.1073/pnas.1816928116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oye K. A., Esvelt K., Appleton E., Catteruccia F., Church G. et al. , 2014. Regulating gene drives. Science 345: 626–628. 10.1126/science.1254287 [DOI] [PubMed] [Google Scholar]
- Port F., Chen H.-M., Lee T., and Bullock S. L., 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 111: E2967–E2976. 10.1073/pnas.1405500111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston C. R., Flores C. C., and Engels W. R., 2006. Differential usage of alternative pathways of double-strand break repair in Drosophila. Genetics 172: 1055–1068. 10.1534/genetics.105.050138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffman E. E., Styhler S., Rother K., Li W., Richard S. et al. , 1998. Premature translation of oskar in oocytes lacking the RNA-binding protein bicaudal-C. Mol. Cell. Biol. 18: 4855–4862. 10.1128/MCB.18.8.4855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H., Nakamura A., and Kobayashi S., 2002. Identification of a transcriptional regulatory region for germline-specific expression of vasa gene in Drosophila melanogaster. Mech. Dev. 112: 129–139. 10.1016/S0925-4773(01)00654-2 [DOI] [PubMed] [Google Scholar]
- Simões M. L., Caragata E. P., and Dimopoulos G., 2018. Diverse Host and Restriction Factors Regulate Mosquito–Pathogen Interactions. Trends Parasitol. 34: 603–616. 10.1016/j.pt.2018.04.011 [DOI] [PubMed] [Google Scholar]
- Van Doren M., Williamson A. L., and Lehmann R., 1998. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8: 243–246. 10.1016/S0960-9822(98)70091-0 [DOI] [PubMed] [Google Scholar]
- Valderrama J. A., Kulkarni S. S., Nizet V., and Bier E., 2019. A bacterial gene-drive system efficiently edits and inactivates a high copy number antibiotic resistance locus. Nat. Commun. 10: 5726 10.1038/s41467-019-13649-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J. H., Nur U., and Wu C.-I., 1988. Selfish genetic elements. Trends Ecol. Evol. 3: 297–302. 10.1016/0169-5347(88)90105-X [DOI] [PubMed] [Google Scholar]
- Werren J. H., 2011. Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc. Natl. Acad. Sci. USA 108: 10863–10870. 10.1073/pnas.1102343108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windbichler N., Papathanos P. A., and Crisanti A., 2008. Targeting the X Chromosome during Spermatogenesis Induces Y Chromosome Transmission Ratio Distortion and Early Dominant Embryo Lethality in Anopheles gambiae. PLoS Genet. 4: e1000291 10.1371/journal.pgen.1000291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windbichler N., Menichelli M., Papathanos P. A., Thyme S. B., Li H. et al. , 2011. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature 473: 212–215. 10.1038/nature09937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., and Finnigan G. C., 2018. Development of a multi-locus CRISPR gene drive system in budding yeast. Sci. Rep. 8: 17277 10.1038/s41598-018-34909-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data that are represented fully within the tables and figures. The nos-, vas-, Ubi-Cas9 plasmids and the corresponding fly lines are deposited at Bloomington Drosophila Stock Center (#79004 – #79006) and AddGene.org (#112685 – #112687), respectively. The BicC-Cas9 and GDe plasmids and fly lines will be made available upon request. Supplemental material available at figshare: https://doi.org/10.25387/g3.11449542.