Abstract
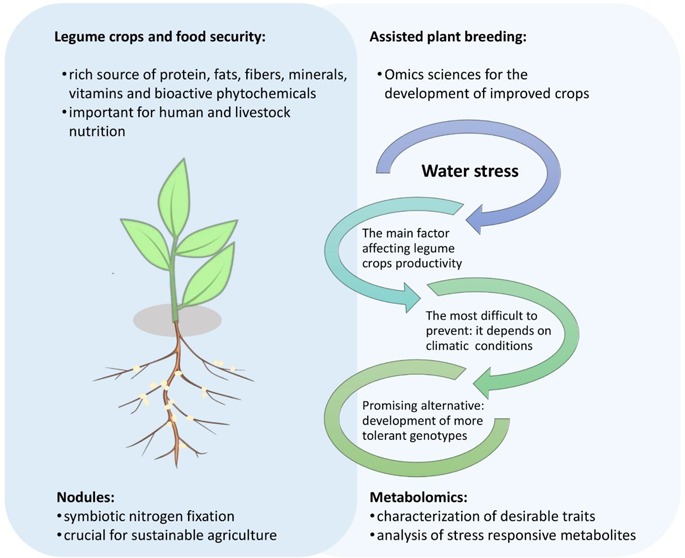
Legume species are an important source of protein and other nutrients for human and livestock consumption, playing a central role in food security. Besides, legumes benefit agriculture because of their ability to establish symbiotic interactions with nitrogen-fixing bacteria, providing nitrogen for subsequent crops, which is very much appreciated for sustainable agricultural practices. However, like other food crops, legumes are highly vulnerable to climate variations, water stresses being the main constraint that negatively affects both crop quality and productivity. Because of this, the development of strategies to improve the tolerance of such cultivars against water stresses, as well as the study of effective approaches to monitor these improvements, have gained special attention during the last years. Among these strategies, metabolomics has been considered one of the most promising approaches for the detection and/or quantification of primary and secondary stress-responsive metabolites in abiotic stresses. In plant science, many research groups have been using metabolomics to evaluate the success of genetic modifications by the analysis of chemical markers that can be altered in breeding programs. In addition, metabolomics is a powerful tool for the evaluation and selection of wild specimens with desirable traits that can be used in the development of improved new cultivars. Therefore, the aim of the present paper is to review the recent progress made in the field of metabolomics and plant breeding, especially concerning the adaptive responses of legume species to abiotic stresses as well as to point out the key primary and secondary metabolites involved in the adaptation and sensing mechanisms.
1. Introduction
One of the major challenges of the modern world is to deal with the sustainable production of food to feed an increasing population. To meet the global demand for food and nutrition, the Food and Agriculture Organization (FAO) estimates that crop productivity must double by 2050. However, due to climate and environmental stress constraints, the productivity scenario of available crops is not promising. By 2080, the productivity of many crops is expected to decrease around 50% in many parts of the world.1,2
The pressures on our current food system include the combination of nonclimate stressors, such as population growth and demand for animal-sourced products and climate change. Climate change is characterized by increased CO2 concentration in the atmosphere, increasing average temperature, and more frequent extreme events including drought periods, heat waves, and flooding. The unpredictable and dynamic nature of changing global climatic conditions impacts food security and, consequently, human health by two main different routes: (i) by affecting the amount of food through direct impact on crop yields and indirectly by impacts on water availability and quality, pests, and disease and pollination services and (ii) by changing CO2 concentration in the atmosphere, affecting plant biomass and nutritional quality.1
Although both biotic and abiotic stressors are challenging for the sustainability of crop production, the agricultural productivity of crops such as wheat, soybean, maize, and potato is more negatively impacted due to abiotic stresses, when compared to biotic agents. Indeed, common climate-related factors, such as water, temperature, and soil salinity, cause significant loss of crop productivity throughout the world.3
For that reason, the development of sustainable agricultural practices, as well as more productive and stress-resistant crop varieties containing genetic traits associated with environmental change adaptation (or greater tolerance to abiotic stresses), will either alone or together be essential to sustainably grow high-yielding crops under increasingly stressful environmental conditions.4
2. Legume Crops and Food Security
The legume family (Fabaceae or Leguminosae) is the third-largest flowering family with over 20 000 species and 750 genera distributed around the globe.5,6 Traditionally, the family is classified into three subfamilies (Caesalpinioideae, Mimosoideae, and Papilionoideae). However, a recent taxonomic revision provided the division of the legume family into six subfamilies (Caesalpinioideae, Cercidoideae, Detarioideae, Duparquetioideae, Dialioideae, and Papilionoideae).2 The family is characterized by its ability to fix nitrogen due to symbiotic nitrogen-fixing associations with bacteria, which is very much appreciated for sustainable crop systems since it provides nitrogen for subsequent crops, with the potential to minimize future nitrogen fertilizer usage.4,5
After the cereals, legumes are the most agriculturally important crop family, playing a central role in food security. Unlike other crops, these species are a rich source of plant-based protein. From the subfamily Papilionoideae, important grain and forage legume species are recognized, mainly because of their economical and nutritional importance.6 Soybean (Glycine max L. Merr.), a warm-season representative, is one of the most important food crops in the world since the grains are a primary and rich source of vegetable oil and protein for human and animal consumption.5
Other warm-season crops such as common bean (Phaseolus vulgaris L.), cowpea (Vigna unguiculata L. Walp), and several Lupines (Lupinus spp. L.), together with cool-season legumes such as lentil (Lens culinaris Medik), chickpea (Cicer arietnum L.), and fava bean (Vicia faba L.), play an important role in the context of food and nutrition security, poverty alleviation, and agricultural sustainability.6 In addition to other legume species, they are also referred to as pulses. Pulses are defined as the dried and edible seeds of certain Leguminosae plants such as dry peas, lentils, beans, and chickpeas. They are characterized by a very high content of protein and fiber and are low in fat. Forage crops such as alfafa (Medicago sativa L.) and Lotus japonicus (Regel) K. Larsen (which are very well-studied model plants) are not considered pulses. Likewise, soybeans and peanuts, which present much higher fat content (oil crops), are not considered pulses either.7
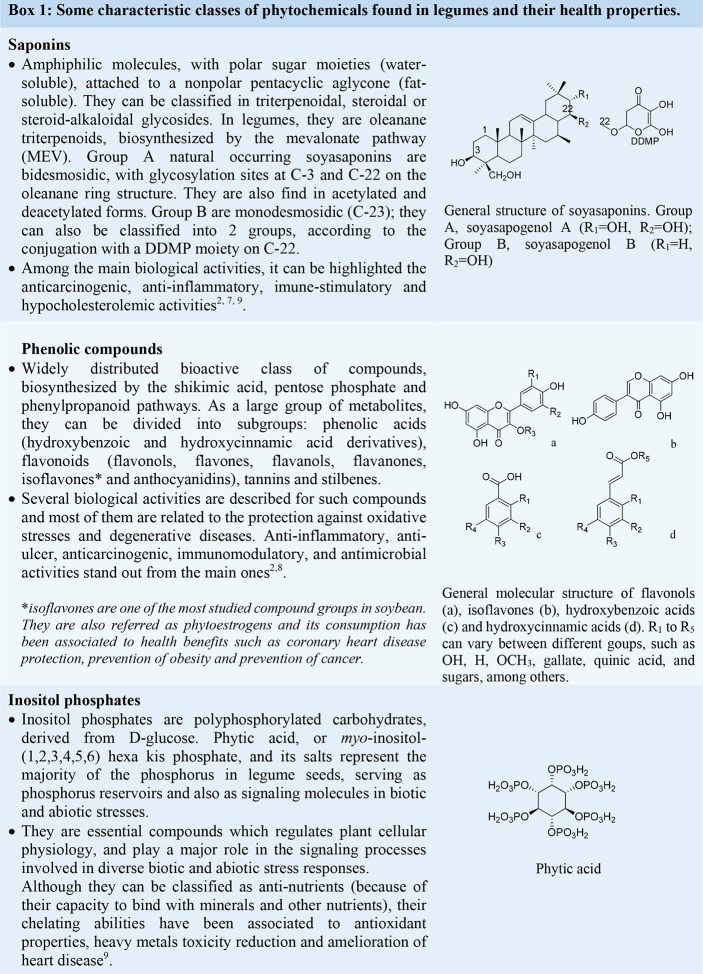
In general, besides proteins and fats, grain legumes are also rich sources of carbohydrates, dietary fibers, vitamins, and minerals, which are essential for both livestock feed and human nutrition.4,5,7,8 From the phytochemical point of view, legumes show a unique pattern compared to other plant species. Besides polyphenols and flavonoids, which are widespread compounds in the plant kingdom, legumes are a rich source of health-promoting compounds such as isoflavones, triterpenoid saponins, and inositol phosphates, among others (Chart 1).2,7,9 Interestingly, during the domestication process of some of these species, the selection of some agricultural traits resulted in an unintended accumulation of some of these compounds (such as some phenolics, flavonoids, and saponins), which also confers resilience to environmental stresses.2
Chart 1.
3. Assisted Breeding for the Development of New Cultivars
Essentially, the natural genetic variation observed in modern cultures, including both intraspecific and interspecific variability, is the result of spontaneous mutations and subsequent selection during the evolution of ancestral forms of plants. After many thousands of years of successive selection and cultivation improvements (considering both domestication and breeding activities), the species that compose our current food system retained only a small part of their original genetic variation. That being said, the wild relatives of such cultures, including those conserved in germplasm collections, are essential resources to retrieve the genetic information lost along the centuries. As repositories of ancestral genes, with rich genetic diversity, they conserve a plethora of desirable traits, such as greater resistance to climate changes and better adaptation to biotic and abiotic stresses.11
With this regard, the development of genetically engineered technologies, such as Agrobacterium-mediated transformation or genome-editing technologies, such as clustered regularly interspaced short palindromic repeats (CRISPR/Cas), zinc-finger nucleases (ZFNs), and transcription activator-like effector nucleases (TALENS), has provided precise and efficient introduction of desirable traits in crop improvements.10 Concomitantly, several technical advances in plant breeding and related techniques also allowed the development of approaches capable of elucidating the associations between genetic and phenotypic variations in plants as well as the identification of molecular markers related with the desirable and/or acquired traits.11
In the last decades, the combination between genomics, transcriptomics, proteomics, and metabolomics has been used to enhance the studies in plant breeding, e.g., to explore processes such as epigenetic regulation, post-transcriptional and post-translational modifications, and other molecular interactions concerned with the genotype–phenotype relationship. Genome-wide association studies (GWASs) on metabolic traits (mGWASs) and metabolomic quantitative trait locus (mQTL) mapping, for example, are modern tools used to link the metabolic phenotypes with the genetic background observed for many complex traits, especially those associated with abiotic stress tolerance.10,12
Metabolomics-assisted breeding, together with other omics, are invaluable tools to facilitate the selection and evaluation of the efficiency of insertion of relevant and desirable metabolic traits, as required by the modern breeding processes. Targeted metabolic profiling, for example, can be used for the identification of plant varieties or individuals that biosynthesize desirable levels of nutraceuticals or secondary metabolites associated with health benefits. The same approach can be used for the identification of molecular markers associated with putative genes and proteins overexpressed by the impact of environmental stressors such as light, temperature, water, or biological agents. Finally, it can be a powerful resource to guide breeding programs, alerting researchers to positive or detrimental traits, and to assist in the elucidation of the regulatory mechanisms.10
In this context, several studies have been reported concerning metabolomics as a tool to explore different aspects in plant breeding, the regulatory mechanisms related to plant growth and development (including those related to crop productivity and performance), adaptation to biotic and abiotic stresses, nutritional improvement, and selection of cultivars for agriculture. Kumar and collaborators (2017)13 reviewed the most recent research and achievements made in the field. For cereal crops, different varieties of maize, rice, potato, and wheat had their metabolites associated with plant tolerance against abiotic stresses, plant signaling, or enhanced nutritional composition. The quality and taste of fruits as well as the relationship between chemical composition and health benefits were also reviewed. Different cultivars of apple, orange, tomato, strawberry, and grape, among others, have been studied and selected for further breeding. The invaluable information includes sugar concentration and diversification, as well as the presence of markers related to ripening, postharvest modifications, and plant infection. Also, information regarding vitamins, phenolic compounds, volatiles, and organic acids, most of them related to taste, quality, and nutritional benefits, has been used for the analysis and selection of cultivars.
4. Metabolomics Approaches for the Study of Crops
Similarly to genomics, proteomics, and transcriptomics (which are approaches used for the high-throughput study of genes, proteins, and transcripts, respectively), metabolomics is a multidisciplinary field of research related to the study of metabolomes of biological systems (e.g., a single cell, a tissue, or an organ, in a given physiological or developmental state).12
The metabolome comprises a pool of low-molecular-weight primary and secondary metabolites (usually <1500 Da), including their precursors and intermediates of the corresponding biosynthetic pathways. Such compounds are considered the end products of gene expression and protein activity, modulating processes between the genome and environment and indicating the functional status of the organism. Moreover, they are an indispensable part of the plant metabolism, influencing all biological processes, such as plant biomass and architecture, and those involved in plant defense or adaptation to biotic and abiotic stresses.12,14
As a high-throughput technology, metabolomics has the fundamental goal to provide a comprehensive view of all metabolites participating in the cellular processes, which requires the use of nonselective, universally applicable, and comprehensive analytical approaches for the identification and quantitation of metabolites. However, due to the enormous existing chemical diversity in nature, there is no single analytical method or protocol capable of furnishing a complete picture of the whole metabolome of an organism.12,15
The plant kingdom may contain from 200 000 to 1 000 000 distinct metabolites, which may be present in a wide range of concentrations (estimated to span 12 orders of magnitude) and vary according to their chemical structures, classes, physicochemical properties, and polarities.15 These particularities make the extraction, measurement, and identification (or even the putative annotation) of such compounds a challenge. For that reason, the identification of both major and minor compounds presented in a plant extract is a complex task. The metabolite profiling methods have to be able to separate and detect a large dynamic range of metabolite classes and concentrations and generate enough spectral information to allow their full or partial identification.12
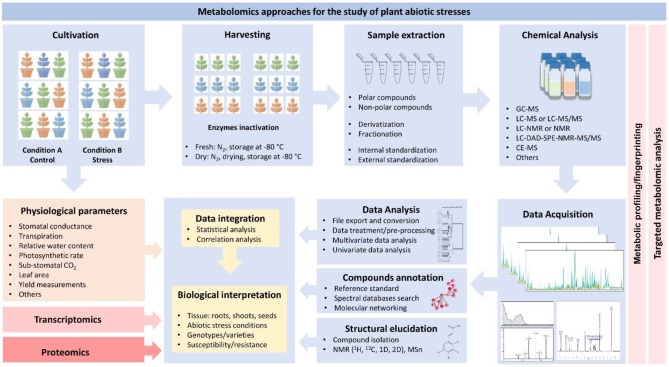
With this regard, the research involving metabolomics and the study of plant species subjected or better adapted to abiotic stresses require different expertise and multidisciplinary knowledge. To make it possible, different protocols and approaches are demanded to accomplish the intended study. It includes the best design of cultivation experiments (under controlled or field conditions), the standardization of robust protocols for sample harvesting, extraction, and chemical analysis, and the establishment of the most suitable workflow for data analysis, data integration (with other metadata or omics results, for example), and biological interpretation of results (Figure 1).
Figure 1.
Schematic representation of the suggested experimental workflow for the metabolomics-assisted study of crops and abiotic stresses. The process starts with the cultivation experiments, which must include at least two different conditions (e.g., stress and control) and a representative number of biological replicates. Depending on the study, different genotypes, varieties, or mutants, susceptible or tolerant, can be arranged and exposed to the experimental conditions. As pointed out by Sanchez and collaborators (2012),19 more than two tolerant and sensitive species/cultivars should be included to avoid a misunderstanding between natural variation and metabolic tolerance. During this phase, the physiological parameters can be monitored and registered. The next step is the harvesting. The plant material (shoots, roots, seed, flowers, stems, or others) is harvested and promptly frozen in liquid nitrogen to avoid enzymatic reactions and degradations. In the sequence, the samples can be stored in a freezer at −80 °C, dried (usually freeze-dried), or directly extracted from the fresh tissue. Before extraction, the samples must be powdered, homogenized, and weighted. The best extraction protocol must be chosen according to the desired purpose (for example, considering targeted metabolomics analysis or metabolic profiling/fingerprinting) and also considering the different classes of metabolites that can be extracted. Usually, internal standardization is required for subsequent normalizations and data analysis. Then, samples are subjected to the chemical analysis (using different analytical platforms). In general, most of the metabolomics protocols include a separation step (by LC or GC, mainly) hyphenated to the detection technique of choice (usually MS or NMR in different arrays). After data acquisition, the raw files are exported for data analysis. The high-throughput process considers several steps such as the conversion to suitable formats, preprocessing, normalizations, data cleaning, alignment, and corrections, among others. Multivariate data analysis methods can be used to evaluate the quality of the acquired data. Additionally, compounds can be annotated by comparing the obtained spectra with those available in mass spectral reference libraries. Still, if necessary, the compounds can be identified by complete structural elucidation (which requires, most of the time, isolation and purification). During this process, the information can be analyzed by different statistical, univariate, or multivariate data analysis tools. Finally, the metabolomics results can be integrated with transcriptomics or proteomics data and/or with the corresponding physiological data for biological interpretation.
By untargeted metabolomic analysis, such as metabolomic profiling and metabolomic fingerprinting, the simultaneous measurement of a large number of metabolites from each sample can be achieved, without establishing a prior specific hypothesis on a particular set of metabolites. As a result, the analysis of the global metabolome expression is performed. Consequently, a large quantity of data is generated, requiring bioinformatics tools to assist in data analysis. In an opposite way, targeted metabolomic analysis is characterized by the measurement of predefined sets of metabolites with a higher level of precision and accuracy.12,16
For that, several analytical separation and detection techniques are usually combined to visualize the metabolome of an organism. Currently, the two main analytical techniques used for the generation of metabolic spectral data are nuclear magnetic resonance (NMR) and mass spectrometry (MS). In the same way, state-of-the-art separation techniques such as gas chromatography (GC), liquid chromatography (LC), and capillary electrophoresis (CE) allow the separation of hundreds of compounds in a single analysis, which is very much appreciated due to the high sample complexity. The hyphenation between these high-resolution separation techniques and accurate tandem mass spectrometry (such as LC-MS/MS, UHPLC-TOF-MS, and CE-MS/MS), or with nuclear magnetic resonance (LC-NMR or LC-DAD-SPE-NMR-MS/MS), together with bioinformatics tools, enabled great advances in the study of plant natural products, providing a more comprehensive view of the metabolome.12,17 In general, LC and GC coupled to accurate tandem mass spectrometry (LC-MS/MS and GC-MS) are the most employed metabolomics platforms due to their unparalleled sensitivity; combined, they also provide an extensive metabolite coverage, which is very much appreciated in metabolomics studies.17 The literature has innumerous papers bringing detailed information and application of such analytical techniques in metabolomics. For that reason, the most common ones will be briefly explained in the following sections.
Gas Chromatography–Mass Spectrometry
Gas chromatography–mass spectrometry (GC-MS) stands out in the analysis of volatile and thermally stable compounds and primary metabolites, including sugars, amino acids, organic acids, and polyamines, as well as biosynthetic pathway precursors. The polar compounds extracted from the plant tissues are first subjected to a derivatization step, making them volatile for further separation into the GC capillary column. Usually, it consists of a two-step derivatization process, which involves a methoxyamination reaction in the carbonyl groups, followed by silylation with N-methyl-trimethylsilyltrifluoroacetamide (MSTFA) or N-O-bistrimethylsilyltrifluoroacetamide (BSTFA), under anhydrous conditions.18 Afterward, samples are subjected to a separation step (using capillary columns, which allow high chromatographic resolving power) and then to the electron impact (EI) fragmentation (a hard ionization method, in which the energy typically applied is set at 70 eV). This robust interface between GC and MS results in a very reproducible fragmentation pattern, which allows the comparison of results with mass spectral databases, boosting peak annotation and compound identification. Among the several metabolite databases available, curated compound reference libraries such as NIST and the Golm Metabolome Database (GMD) stand out for metabolomics studies. Concerning mass detection, time-of-flight (TOF-MS) has become the method of choice because of advantages such as fast scan times, improved deconvolution, and high mass accuracy.15 However, a major advantage of GC-TOF-MS systems is their enhanced software capability, which supports automated and comprehensive extraction of all mass spectra from a chromatogram, in-built mass-spectral correction for coeluting metabolites, calculation of retention-time indices, and automated picking of a suitable fragment mass for selective quantification. Moreover, GC-TOF-MS has the potential to be truly nonbiased and fully automated with respect to metabolite identification, although it still requires expert inputs to correct inappropriate assignments.18
Liquid Chromatography–Mass Spectrometry
Despite the outstanding GC-MS capacity to separate, resolve, and detect a wide range of metabolites from complex matrices, one of its limitations is the inability to ionize thermolabile or high molecular mass metabolites. For that reason, LC-MS is the technique of choice for the analysis of thermolabile unstable compounds, polar and relatively nonpolar compounds, and mainly secondary metabolites and phytohormones, without any derivatization step. Liquid chromatography–mass spectrometry is a very versatile analytical platform and can be found in different arrays. A plethora of available chromatographic columns, differing in length, diameter, resolution, and stationary phases, together with a vast combination of mobile phases, allow the separation of the plant natural compounds according to their chemical properties.15 Electrospray ionization (ESI), a soft-ionization method, is the most used technique since it allows the direct analysis of compounds from the liquid phase. Detailed structural information can be achieved by collision-induced dissociation (CID), which is generally carried out on a tandem MS instrument (allowing tandem MSn experiments).16 After the ionization step, the fragments are driven to the analyzer. Different types of analyzers are commercially available. It includes (i) tandem-in-time instruments, such as quadrupole ion traps (QIT-MS), orbitrap, and Fourier transform ion cyclotron resonance (FT-ICR)-MS and (ii) tandem-in-space instruments, such as triple quadrupoles (QqQ) and quadrupole time-of-flight (qTOF). The choice depends on the sensitivity, mass resolution, and dynamic range required.15
Nuclear Magnetic Resonance
Nuclear magnetic resonance (NMR) is a highly reproducible and universal (nonselective) spectroscopic technique. The resonances recorded in an NMR spectrum provide compound identification through the interpretation of the chemical shifts and coupling constants. Moreover, it can differentiate compounds with identical masses but different spatial configuration. In addition, the spectral peak areas generated by the signals can be used for the determination of compound concentration, which provides an advantage over the MS-based techniques due to its higher accuracy and reproducibility. In general, the main compounds covered by NMR in metabolome analysis are carbohydrates, amino acids, and organic acids. However, the sensitivity of NMR is much lower than that of MS-based techniques, which restrains its application in the analysis of low-abundant plant metabolites. While 1D NMR (mainly 1H NMR) is the most commonly used method in high-throughput metabolomics studies, 2D NMR is often used for the characterization of those compounds that cannot be identified with 1D NMR spectra. For the structural elucidation of isolated natural product compounds, further NMR experiments may include the acquisition of 13C NMR data, as well as (1H,1H)-COSY, (1H,1H)- TOCSY, (1H,1H)-NOESY, (1H,1H)-ROESY, (1H,13C)-HSQC, and (1H,13C)-HMBC, among others.12
Analysis and Interpretation of the Metabolomic Data
The high-throughput metabolite profiling strategies applied in metabolomics generate a great amount of raw data, which must be extracted, converted, deconvoluted, aligned, and organized into a matrix size of N × M (where N is the number of biological and technical replicates and M is the metabolites or signals). Afterward, and prior to the statistical analysis, it is often necessary to perform additional data preprocessing steps such as normalizations, noise filtering, baseline correction, smoothing, scaling, replacement of missing values, elimination of outliers, and data reduction, if necessary.16
Typically, both multivariate and univariate methods are used for the analysis of the metabolomics data. Multivariate statistical methods, including unsupervised principal component analysis (PCA) or supervised partial least-squares (PLS, OPLS, PLS-DA, etc.), allow exploration and visualization of metabolomics datasets through simultaneous analysis of multiple variables. Univariate methods, such as ANOVA, Student’s, Tukey, and Welch’s t-test, are especially interesting for the precise identification of down- and upregulated metabolic traits. If the information concerning the identity of the detected compounds is not available, the analysis can be done considering specific classes of metabolites.10
Indeed, only a small fraction of the detected metabolites can be identified, which requires, most of the time, the isolation and acquisition of unambiguous results. Regarding any technique of choice, metabolite identification is one of the bottlenecks in metabolomics.10,16 Depending on the instrumentation, availability of reference standards, and consistent databases, only the partial assignments or putative identification is possible. In this case, compounds are annotated and not always identified.
However, different levels of confidence of metabolite annotation can be achieved, ranging from Level 0 (or identified compound, including full 3D structure and stereochemistry), Level 1 (the most common, achieved by two orthogonal parameters, such as retention time and MS/MS spectrum), Levels 2 and 3 (putatively annotated compounds and compound classes; accurate mass measurement and isotope abundance ratios are also possible), to Level 4 (unidentified or unclassified metabolites that can be differentiated based on analytical data).17
In general, the different levels of annotations can be achieved with the assistance of MS- (or NMR-) based spectral databases and in silico spectral prediction algorithms. Among the most important databases, the following can be highlighted: NIST (https://www.nist.gov), Wiley (https://www.sisweb.com/software/ms/wiley.htm), MassBank (https://massbank.eu/MassBank/), GMD (http://gmd.mpimp-golm.mpg.de/), MoNA (https://mona.fiehnlab.ucdavis.edu), and METLIN (https://metlin.scripps.edu), which contain tens of thousands of verified experimental mass spectra, and the Global Natural Products Social Molecular Networking - GNPS database (https://gnps.ucsd.edu), which also allows us to upload and share experimental spectra.10,16,17
In silico prediction algorithms assist compound annotation by predicting the most likely chemical structure that corresponds to a given experimental mass spectrum, using the information obtained from known compounds stored in chemical databases.10 For that, four general methods can be distinguished (quantum chemistry, machine learning, heuristic-based methods, and chemical-reaction-based methods), and the available algorithms use EI-MS and/or CID-MS/MS data.10,17
Finally, it is important to point out that even if the compound annotation or identification is not possible the untargeted metabolomics results are still able to reveal up- and down-regulated metabolites in a given sample group relative to controls. Depending on the results, compound identification can be carried out by traditional phytochemical methods, including the isolation, purification, and complete structural elucidation.10,16
5. Plant Responses to Environmental Stresses
Many responses to environmental stresses such as excessive light, water stresses, salinity, extreme temperatures, nutrient deficiency, metal toxicity, or UV–B radiation are common among plants. To counteract the adverse environmental conditions, plants need to develop mechanisms for adaptation and acclimatization, which requires a new state of cellular homeostasis. While adaptation is related to an evolutionary process (considering populations over many generations), acclimation involves reprogramming of their development, physiology, and metabolism, enabling their survival.19 Abiotic stresses are directly related to changes in several cellular metabolic pathways, such as carbohydrates, amino acid, and peptide metabolism. However, if the metabolic homeostasis is disrupted, there might be a reduction in growth, development, and, consequently, yield.14
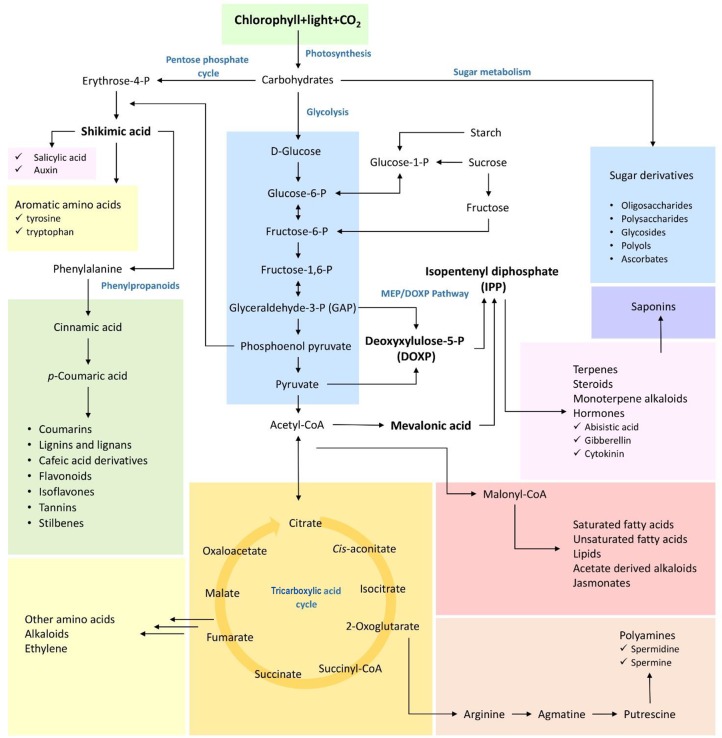
The alterations in plant metabolism are an important part of early stress responses and occur in all plant species. They include the accumulation of compatible solutes for the retention of water in the cell and antioxidants for protection against the oxidative damage to cellular components, for example. During a drought, soluble compounds, such as oligosaccharides, polyhydric alcohols, sucrose, mannose, trehalose, amino acids, and polyamines, assist with maintaining turgor, membrane integrity, and stabilizing macromolecules,20 thus contributing to keeping cell homeostasis. Obata and Fernie15 reviewed and compared metabolic fingerprints of Arabidopsis leaves when subjected to stresses such as dehydration, salt, temperature, light, ultraviolet (UV) light, and low nitrogen amounts, among others. They reported that specific compounds such as sucrose, raffinose, proline, other branched chain amino acids, and γ-aminobutyric acid (GABA) are generally accumulated in different levels during most of the abiotic stresses, showing that the stress-specific plant response is the result of an inhibition or activation of a defined metabolic pathway (Figure 2).
Figure 2.
Schematic representation of plant biosynthetic pathways involved in the biosynthesis of primary and secondary metabolites in plants. Some of the represented compounds and precursors have their concentrations altered in response to biotic and abiotic stresses. The understanding of the dynamics and mechanisms of such alterations, as well as their biological functions, is fundamental to support the development of new cultivars, more tolerant or resistant to adverse environmental conditions. In this context, metabolic profiling strategies are invaluable tools to access the information concerning these metabolic alterations.
Although all environmental stresses are challenging for plants, a drought is by far the most complex and unpredictable abiotic stress that affects plant growth, development, survival, and crop productivity at a global level. Plants subjected to a drought have essential physiological functions disturbed. In the cellular level, damages in the photosynthetic apparatus, alterations in the carbon assimilation rate, increased oxidative damage, alterations on enzyme activities, and ion imbalance, among others, can be highlighted. Consequently, plants show deficiencies in water, nitrogen and mineral acquisition, and reduced germination. Altogether, these alterations cause considerable yield reduction. From the molecular point of view, in general, water stresses can significantly change both the transcriptomic and metabolic profiles, which include compatible solute accumulation and the production of antioxidants and hormones for physiological regulation, among others.20
6. Metabolic Response of Legumes to Drought
Similarly to other plant species, stressful environments negatively impact legume production, decreasing the yield and quality. Moreover, the biological nitrogen fixation is impaired in dehydrated tissue, which happens when plants are subjected to drought. The consequent yield loss is caused by inadequate oxygen supply, carbon shortage, nitrogen feedback, oxidative stress, and sulfur metabolism modifications.4
Despite the relevance of legumes for agriculture and human health and nutrition, the increases in grain legume yields are around 0–2% per year, which is much lower when compared with cereals. To sustain the production growth, productivity improvements will be necessary, as these recorded production rates are due to increases in the land planted area and not due to the performance of the cutivars.5 Considering this scenario, the productivity could be considerably improved by the introduction of new varieties better adapted to water stresses. For legumes, many wild species grow in arid regions, which make them good models for understanding the tolerance mechanisms. Besides, these wild relatives can furnish novel alleles for crop improvement using various biotechnological approaches. It may include abiotic stress tolerance, biotic stress resistance, improvement on yield, and better nutritional composition, among others.2
One of the mechanisms described for plant drought stress tolerance is the production and accumulation of organic solutes. These compounds may act as osmolytes for the osmostic adjustment and turgor maintenance in the cytosol or as osmoprotectants to stabilize cellular components. Therefore, the regulation of these compounds constitutes one of the most promising alternatives for the improvement of drought tolerance in legume crops. Consequently, a deeper understanding about the metabolic pathways that produce commonly regulated osmolytes is essential for the success of the plant breeding approaches.19,21
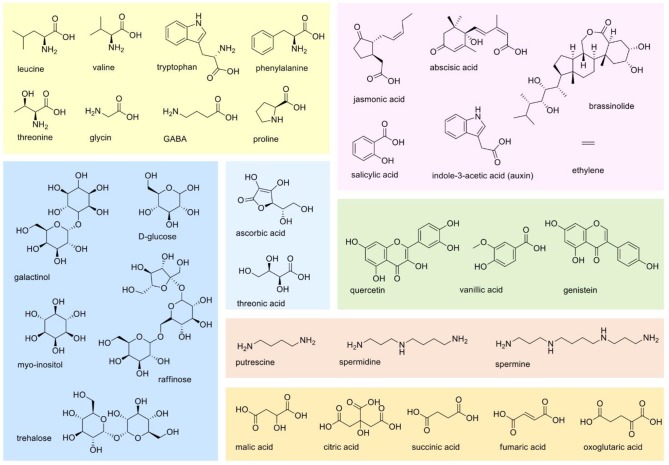
Given the importance for a better understanding about how legumes respond and adapt to drought conditions, the metabolic responses of legume species have been reported in several studies. In these and in others involving various plant species and crops, some key primary and secondary metabolites have been associated with the drought responses (Chart 2, Figure 3). For that, different analytical platforms and design of experiments have been used.
Chart 2.
Figure 3.
Primary and secondary metabolites that had their concentrations altered in legume species subjected to water stresses (as pointed out in Chart 2). Representative compounds were organized according to their biosynthetic pathways as shown in Figure 2. In blue, sugars; in light blue, antioxidants; in light orange, polyamines; in bright yellow, amino acids; in pink, phytohormones; in green, phenolic compounds; in yellow, tricarboxylic acid TCA-derived compounds.
Goufo and collaborators21 used GC-TOF-MS and LC-DAD for the metabolomic profiling of cowpea (Vigna unguiculata L. Walp.) when subjected to drought (for 6 or 12 days) and rewatered for 6 days after 6 days of stress. Although cowpea is considered a drought-resistant crop for presenting desirable physiological traits, drought still impacts its productivity. By GC-MS, the authors were able to identify 41 primary metabolites, including 5 sugars, 4 polyols, 24 amino acids and/or their derivatives, and 8 organic acids. By LC-DAD, 35 peaks were assigned based on their spectral features. It included the annotation of 15 phenolic acids, 17 flavonoids, and 3 proanthocyanidins. Therefore, by coupling two complementary analytical platforms, not only primary metabolites were assigned as resistance markers or part of an adaptive response to drought stress but also secondary metabolites were indicated. Quercetin 3-O-6″-malonylglycoside, kaempferol 3-O-diglycoside, quercetin, galactinol, and proline were identified as having the most significant responses to drought.
Coutinho and collaborators22 analyzed the leaves and roots from two different soybean cultivars (one sensitive and one moderately tolerant) by 1H NMR and multivariate data analysis. The analysis of the chemical shifts and coupling constants allowed them to assign the signals of 26 primary and 11 secondary metabolites. The authors showed that alanine, GABA, sucrose, acetate, citrate, and succinate were accumulated in the roots when plants were subjected to flooding. Interestingly, most of the levels of these compounds decreased in the leaves.
The differential accumulation of metabolites was also described for two varieties of chickpea (Cicer arietinum L.), one tolerant and one sensitive, both exposed to drought stress. The UPLC-HRMS-based untargeted metabolic profiling approach allowed the separation of 691 peaks, from which 175 were identified as known metabolites, including amino acids, organic acids, sugars, polyamines, nitrogenous compounds, and polyphenols. Proline, arginine, histitine, isoleucine, and tryptophan were highly accumulated in the leaves of the tolerant variety after drought stress induction, while compounds such as GABA, alanine, α-ketoglutaric acid, choline, phenylalanine, tyrosine, glucosamine, adenosine, guanine, and aspartic acid were decreased in the two varieties.23
In another study, the nodules of two varieties of peanut (Arachis hypogaea L.) with contrasting tolerance to drought (one sensitive and one more tolerant) were analyzed by GC-MS after being subjected to drought stress.4 From the 58 identified metabolites, amino acids, organic acids, carbohydrates, and some precursors showed significant changes. The nodules of the tolerant cultivar accumulated trehalose, proline, and GABA, while the sensitive cultivar had the levels of asparagine and glutamine decreased. Besides, this cultivar was unable to recover the metabolism similarly to well-watered plants.
Finally, a study concerning the drought acclimation process in six Lotus species showed that the responses are accompanied by increases in the cellular concentration of many small hydrophilic metabolites (or osmolytes, such as amino acids, sugars, and TCA cycle metabolites), which can vary according to the studied species. In their study, 198 mass spectral tags were manually annotated from the GC-MS profiling data set, representing known and still unknown metabolites. From those, 90 were altered by the drought treatment (76 increased and 14 decreased). Significant increases were observed for organic acids (succinic and malic acid), sugars (fructose, glucose galactose, and maltose) and polyols (arabitol, ononitol, and galactitol). Considering the amino acids, proline, leucine, and isoleucine increased, while serine, glycine, and threonine decreased. Asparagine, lysine, and valine showed no significant change. Finally, the authors point out that robust changes involved in the acclimation across a genus must be analyzed using more than two tolerant versus sensitive species/cultivars to avoid a misunderstanding between natural variation and metabolic tolerance.19
7. Conclusions and Future Research Perspectives
Our current food system is based on a finite number of domesticated species, which were selected over the centuries. Consequently, only part of the natural original genetic variation was retained. Although the domestication processes allowed the selection of the most interesting varieties in terms of yield, the productivity of many crops has already been impacted by the global climate change. Considering the increasing demand for food to support an increasing population, as well as the negative projections concerning climate changes, the development of new cultivars capable of adapting to abiotic events is essential for the protection of plantations, increase of quality and productivity, and reduction of production costs. It is especially important for legume crops, as they constitute one of the most promising crop families for food security, given their significant nutritional value. Moreover, their ability to perform symbiotic nitrogen fixation is crucial for the development of sustainable agricultural practices.
For that reason, in the last decades, many efforts have been made aiming at the development and characterization of more tolerant plant genotypes for agriculture. However, the development of abiotic stress-resilient crops requires a deep knowledge about plant development and the biological processes that enable plants to survive in stressful environments. Genomics and transcriptomics have been extensively used for the development and characterization of more tolerant plant genotypes. More recently, studies concerning plant genetic and epigenetic control, post-transcriptional and post-translational modifications, biochemical interactions, and metabolic changes, among others, have been carried out in order to understand the genotype–phenotype relationship. Modern approaches such as mGWASs and mQTL mapping have been used in plant science to associate metabolic phenotypes with genotypes which show desirable traits for plant breeding.
However, the identification of molecular traits that vary in response to stress events remains a challenge. In this context, metabolic profiling can be used to characterize the molecular traits involved in such processes, providing valuable information to guide the breeding programs (Figure 1). Indeed, metabolomics is a powerful tool to measure the biological or physiological responses to environmental changes, especially when it is integrated with other profiling technologies such as transcriptomics and proteomics.
For that, state-of-the-art analytical technologies and approaches are in constant development. They include: (i) the standardization and validation of protocols for sample extraction and preparation, (ii) improvements in resolution power and sensitivity of equipments, (iii) advancements in the application and integration of multiple analytical platforms (such as NMR, GC-MS, and LC-MS/MS or LC-SPE-NMR-MS), and (iv) developments of algorithms, bioinformatics, and chemoinformatics tools for deconvolution and data analysis, among others. Besides, open-access natural chemical compound databases (derived from both model and nonmodel organisms) have been considered an invaluable resource for the annotation of the metabolites.
To understand the responsible mechanisms that plants use for biotic and abiotic stress adaptation, the metabolomics approaches include the identification and quantitation of the stress-responsive specialized metabolites produced by plants when subjected to adverse envionmental conditions. In water stresses, alterations in the biosynthesis of osmolytes, antioxidants, hormones, polyamines, and other signaling molecules have been described for many species, including both model plants and crops. Although the up- or down-regulation of some compounds is common among many species (e.g., glucose, raffinose, proline, and phytohormones), the mechanisms and dynamics of these alterations are still very much incomplete. Moreover, alterations in the production of some secondary metabolites seem to be specific and related to the genetic background of each species. Interestingly, most of the secondary metabolites reported so far have a close relationship with antioxidant properties. It is interesting to point out that, while changes in the primary metabolism are one of the first responses to several biotic and abiotic stresses, the secondary metabolism is less restricted and could configure the end points for storing the information acquired during the adaptation process. For that reason, we believe that special attention has to be given to the integration between primary and secondary metabolism, which can certainly contribute to the elucidation of the tolerance mechanisms involved in plant adaptations to environmental stresses.
Acknowledgments
The authors acknowledge the support of the São Paulo Research Foundation (FAPESP grants #2017/19702-6 and #2019/08477-7 (PCPB) and #2014/50265-3 (NPL)). This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001 and CNPq (305297/2017-1 and 408931/2016-7). We are also grateful to Dr. Joachim Kopka and Alexander Erban from Max-Planck Institute of Molecular Plant Physiology for the fruitful discussions and support.
Biographies
Paula Carolina Pires Bueno is a postdoctoral research scientist, specializing in Natural Products Chemistry. She has a B.Sc. degree in Pharmaceutical Sciences and Biochemistry (University of São Paulo, Brazil), a M.Sc. degree in Pharmaceutical Sciences, with a concentration area of Synthetic and Natural Products (University of São Paulo, Brazil), and a Ph.D. in Chemistry (São Paulo State University, Brazil). She also conducted part of her activities as a guest researcher at the Institut für Biologie and Biotechnologie der Pflanzen (Westfälische Wilhelms Universität Münster, Germany), at the Institute of Biology of Leiden (University of Leiden, Netherlands), and she is currently a guest postdoctoral researcher at the Max-Planck Institute of Molecular Plant Physiology (MPI-MP, Germany). Her research, conducted in the last 14 years, is focused on the field of plant metabolomics, analytical chemistry, and plant and food chemistry.
Norberto Peporine Lopes obtained a B.Sc. and M.Sc. degree in Pharmacy and a Ph.D. in Chemistry at the University of São Paulo and a Post-Doc at Cambridge University, UK. At present, he is a Full Professor of Organic Chemistry at the University of São Paulo at the School of Pharmaceutical Sciences of Ribeirão Preto – University of São Paulo. Currently, he is President of the Brazilian Chemical Society, Fellow of the Royal Society of Chemistry, Member of the American Chemical Society, Member of GA, and Advisor of the Brazilian Society of Mass Spectrometry. He was elected in 2017 as a member of the Brazilian Academy of Sciences. His research deals with organic chemistry, with emphasis on natural products chemistry and mass spectrometry.
The authors declare no competing financial interest.
References
- Intergovernmental Panel on Climate Change (IPCC) . Climate Change and Land: Summary for Policymakers. An IPCC Special Report on climate change, desertification, land degradation, sustainable land management, food security, and greenhouse gas fluxes in terrestrial ecosystems; 2019. 10.4337/9781784710644. [DOI]
- Zhang H.; Yasmin F.; Song B. H. Neglected treasures in the wild — legume wild relatives in food security and human health. Curr. Opin. Plant Biol. 2019, 49, 17–26. 10.1016/j.pbi.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciura J.; Kruk J. Phytohormones as targets for improving plant productivity and stress tolerance. J. Plant Physiol. 2018, 229, 32–40. 10.1016/j.jplph.2018.06.013. [DOI] [PubMed] [Google Scholar]
- Furlan A. L. F.; Bianucci E.; Castro S.; Dietz K. Metabolic features involved in drought stress tolerance mechanisms in peanut nodules and their contribution to biological nitrogen fixation. Plant Sci. 2017, 263, 12–22. 10.1016/j.plantsci.2017.06.009. [DOI] [PubMed] [Google Scholar]
- Foyer C. H.; Lam H.; Nguyen H. T.; Siddique K. H. M.; Varshney R. K.; Comer T. D.; Cowling W.; Bramley H.; Mori T. A.; Hodgson J. M.; Cooper J. W.; Miller A. J.; Kunert K.; Vorster J.; Cullis C.; Ozga J. A.; Wahlqvist M. L.; Liang Y.; Shou H.; Shi K.; Yu J.; Fodor N.; Kaiser B. N.; Wong F.; Valliyodan B.; Considine M. J. Neglecting legumes has compromised human health and sustainable food production. Nat. Plants 2016, 2, 1–10. 10.1038/nplants.2016.112. [DOI] [PubMed] [Google Scholar]
- Mousavi-Derazmahalleh M.; Bayer P. E.; Hane J. K.; Valliyodan B.; Nguyen H. T.; Nelson M. N.; Erskine W.; Varshney R. K.; Papa R.; Edwards D. Adapting legume crops to climate change using genomic approaches. Plant, Cell Environ. 2019, 42, 6–19. 10.1111/pce.13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B.; Singh J. P.; Singh N.; Kaur A. Saponins in pulses and their health promoting activities: A review. Food Chem. 2017, 233, 540–549. 10.1016/j.foodchem.2017.04.161. [DOI] [PubMed] [Google Scholar]
- Singh B.; Singh J. P.; Kaur A.; Singh N. Phenolic composition and antioxidant potential of grain legume seeds: A review. Food Res. Int. 2017, 101, 1–16. 10.1016/j.foodres.2017.09.026. [DOI] [PubMed] [Google Scholar]
- Muzquiz M.; Varela A.; Burbano C.; Cuadrado C.; Guillamon E.; Pedrosa M. M. Bioactive compounds in legumes: Pronutritive and antinutritive actions, implications for nutrition and health. Phytochem. Rev. 2012, 11, 227–244. 10.1007/s11101-012-9233-9. [DOI] [Google Scholar]
- Christ B.; Pluskal T.; Aubry S.; Weng J. K. Contribution of untargeted metabolomics for future assessment of biotech crops. Trends Plant Sci. 2018, 23, 1047–1056. 10.1016/j.tplants.2018.09.011. [DOI] [PubMed] [Google Scholar]
- Fernie A. R.; Schauer N. Metabolomics-assisted breeding: a viable option for crop improvement?. Trends Genet. 2009, 25, 39–48. 10.1016/j.tig.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Brunetti A. E.; Carnevale Neto F.; Vera M. C.; Taboada C.; Pavarini D. P.; Bauermeister A.; Lopes N. P. An integrative omics perspective for the analysis of chemical signals in ecological interactions. Chem. Soc. Rev. 2018, 47, 1574–1591. 10.1039/C7CS00368D. [DOI] [PubMed] [Google Scholar]
- Kumar R.; Bohra A.; Pandey A. K.; Pandey M. K.; Kumar A. Metabolomics for plant improvement: Status and prospects. Front. Plant Sci. 2017, 8, 1–27. 10.3389/fpls.2017.01302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A.; Rushton P. J.; Rohila J. S. Metabolomic profiling of soybeans (Glycine max L.) reveals the importance of sugar and nitrogen Metabolism under Drought and Heat Stress. Plants 2017, 6, 21. 10.3390/plants6020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata T.; Fernie A. R. The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci. 2012, 69, 3225–3243. 10.1007/s00018-012-1091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksenov A. A.; Da Silva R.; Knight R.; Lopes N. P.; Dorrestein P. C. Global chemical analysis of biology by mass spectrometry. Nat. Rev. Chem. 2017, 1, 1–20. 10.1038/s41570-017-0054. [DOI] [Google Scholar]
- Blaženović I.; Kind T.; Ji J.; Fiehn O. Software tools and approaches for compound identification of LC-MS/MS data in metabolomics. Metabolites 2018, 8, 31. 10.3390/metabo8020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorge T. F.; Rodrigues J. A.; Caldana C.; Schmidt R.; van Dongen J. T.; Thomas-Oates J.; Antonio C. Mass spectrometry-based plant metabolomics: metabolite responses to abiotic stress. Mass Spectrom. Rev. 2016, 35, 620–649. 10.1002/mas.21449. [DOI] [PubMed] [Google Scholar]
- Sanchez D. H.; Schwabe F.; Erban A.; Udvardi M. K.; Kopka J. Comparative metabolomics of drought acclimation in model and forage legumes. Plant, Cell Environ. 2012, 35, 136–149. 10.1111/j.1365-3040.2011.02423.x. [DOI] [PubMed] [Google Scholar]
- Nadeem M.; Li J.; Yahya M.; Sher A.; Ma C.; Wang X.; Qiu L. Research Progress and Perspective on Drought Stress in Legumes: A Review. Int. J. Mol. Sci. 2019, 20, 2541. 10.3390/ijms20102541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goufo P.; Moutinho-Pereira J. M.; Jorge T. F.; Correia C. M.; Oliveira M. T.; Rosa E. A. S.; Antonio C.; Trindade H. Cowpea (Vigna unguiculata L. Walp.) metabolomics: Osmoprotection as a physiological strategy for drought stress resistance and improved yield. Front. Plant Sci. 2017, 8, 1–22. 10.3389/fpls.2017.00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho I. D.; Henning L. M. M.; Dopp S. A.; Nepomuceno A.; Moraes L. A. C.; Marcolino-Gomes J.; Richter C.; Schwalbe H.; Colnago L. A. Flooded soybean metabolomic analysis reveals important primary and secondary metabolites involved in the hypoxia stress response and tolerance. Environ. Exp. Bot. 2018, 153, 176–187. 10.1016/j.envexpbot.2018.05.018. [DOI] [Google Scholar]
- Khan N.; Bano A.; Rahman M. A.; Rathinasabapathi B. UPLC-HRMS-based untargeted metabolic profiling reveals changes in chickpea (Cicer arietinum) metabolome following long-term drought stress. Plant, Cell Environ. 2019, 42, 115–132. 10.1111/pce.13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fàbregas N.; Fernie A. R. The metabolic response to drought. J. Exp. Bot. 2019, 70, 1077–1085. 10.1093/jxb/ery437. [DOI] [PubMed] [Google Scholar]
- Pál M.; Szalai G.; Janda T. Speculation: Polyamines are important in abiotic stress signaling. Plant Sci. 2015, 237, 16–23. 10.1016/j.plantsci.2015.05.003. [DOI] [PubMed] [Google Scholar]