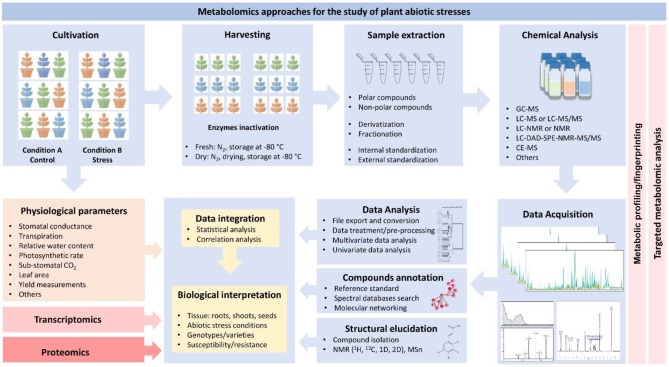
Figure 1.
Schematic representation of the suggested experimental workflow for the metabolomics-assisted study of crops and abiotic stresses. The process starts with the cultivation experiments, which must include at least two different conditions (e.g., stress and control) and a representative number of biological replicates. Depending on the study, different genotypes, varieties, or mutants, susceptible or tolerant, can be arranged and exposed to the experimental conditions. As pointed out by Sanchez and collaborators (2012),19 more than two tolerant and sensitive species/cultivars should be included to avoid a misunderstanding between natural variation and metabolic tolerance. During this phase, the physiological parameters can be monitored and registered. The next step is the harvesting. The plant material (shoots, roots, seed, flowers, stems, or others) is harvested and promptly frozen in liquid nitrogen to avoid enzymatic reactions and degradations. In the sequence, the samples can be stored in a freezer at −80 °C, dried (usually freeze-dried), or directly extracted from the fresh tissue. Before extraction, the samples must be powdered, homogenized, and weighted. The best extraction protocol must be chosen according to the desired purpose (for example, considering targeted metabolomics analysis or metabolic profiling/fingerprinting) and also considering the different classes of metabolites that can be extracted. Usually, internal standardization is required for subsequent normalizations and data analysis. Then, samples are subjected to the chemical analysis (using different analytical platforms). In general, most of the metabolomics protocols include a separation step (by LC or GC, mainly) hyphenated to the detection technique of choice (usually MS or NMR in different arrays). After data acquisition, the raw files are exported for data analysis. The high-throughput process considers several steps such as the conversion to suitable formats, preprocessing, normalizations, data cleaning, alignment, and corrections, among others. Multivariate data analysis methods can be used to evaluate the quality of the acquired data. Additionally, compounds can be annotated by comparing the obtained spectra with those available in mass spectral reference libraries. Still, if necessary, the compounds can be identified by complete structural elucidation (which requires, most of the time, isolation and purification). During this process, the information can be analyzed by different statistical, univariate, or multivariate data analysis tools. Finally, the metabolomics results can be integrated with transcriptomics or proteomics data and/or with the corresponding physiological data for biological interpretation.