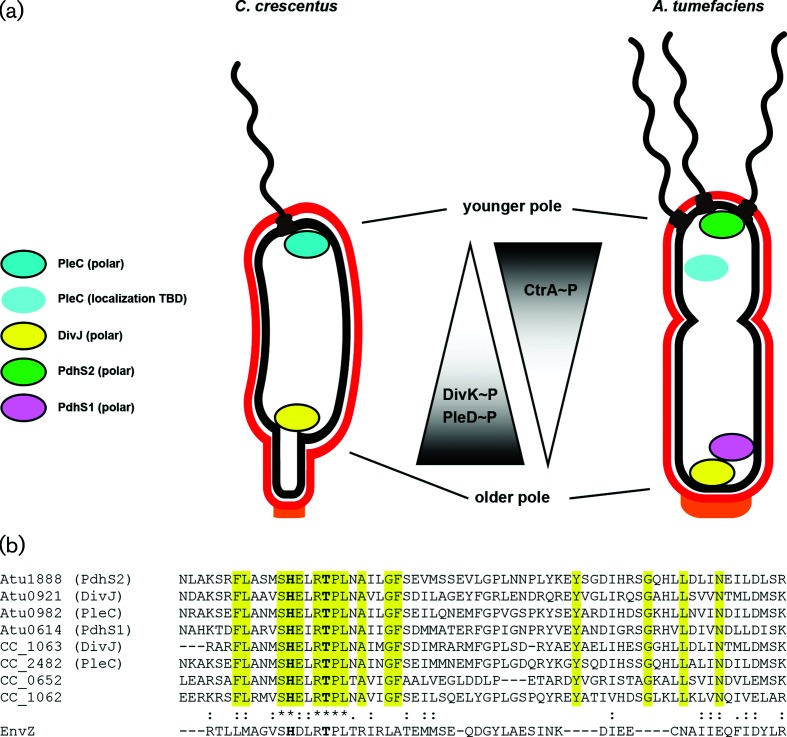
Fig. 1.
The PdhS kinases of C. crescentus and A. tumefaciens localize differentially and affect phenotypic outputs through the response regulators DivK, PleD and CtrA. (a) Cartoon model of known localization of the namesake PdhS kinases from C. crescentus , PleC and DivJ, and three PdhS kinases from A. tumefaciens , PdhS1, PdhS2 and DivJ. Kinases represented as coloured ovals with a black border experimentally localize to the indicated poles. The PleC oval without a border has not been experimentally demonstrated to localize in A. tumefaciens . As a result of this localization, the phosphorylation status of the direct PdhS kinase targets, DivK and PleD, and the indirect target, CtrA, may be differentially affected. (b) Multiple sequence alignment of the HisKA domain from the PdhS kinases of C. crescentus and A. tumefaciens . Sequences were aligned using the Clustal Omega web service hosted by the European Molecular Biology Laboratory (EMBL) – European Bioinformatics Institute. The four PdhS kinases from A. tumefaciens plus PleC and DivJ from C. crescentus were included. Also included were two additional predicted PdhS kinases, CC_0652 and CC_1062, from C. crescentus . The EnvZ sensor kinase is included for comparison. Yellow highlighting indicates residues that define the PdhS kinases. The conserved histidine and threonine residues mutated in this work are in bold.