Figure 3. Structure of the EHEC PpdD pilus.
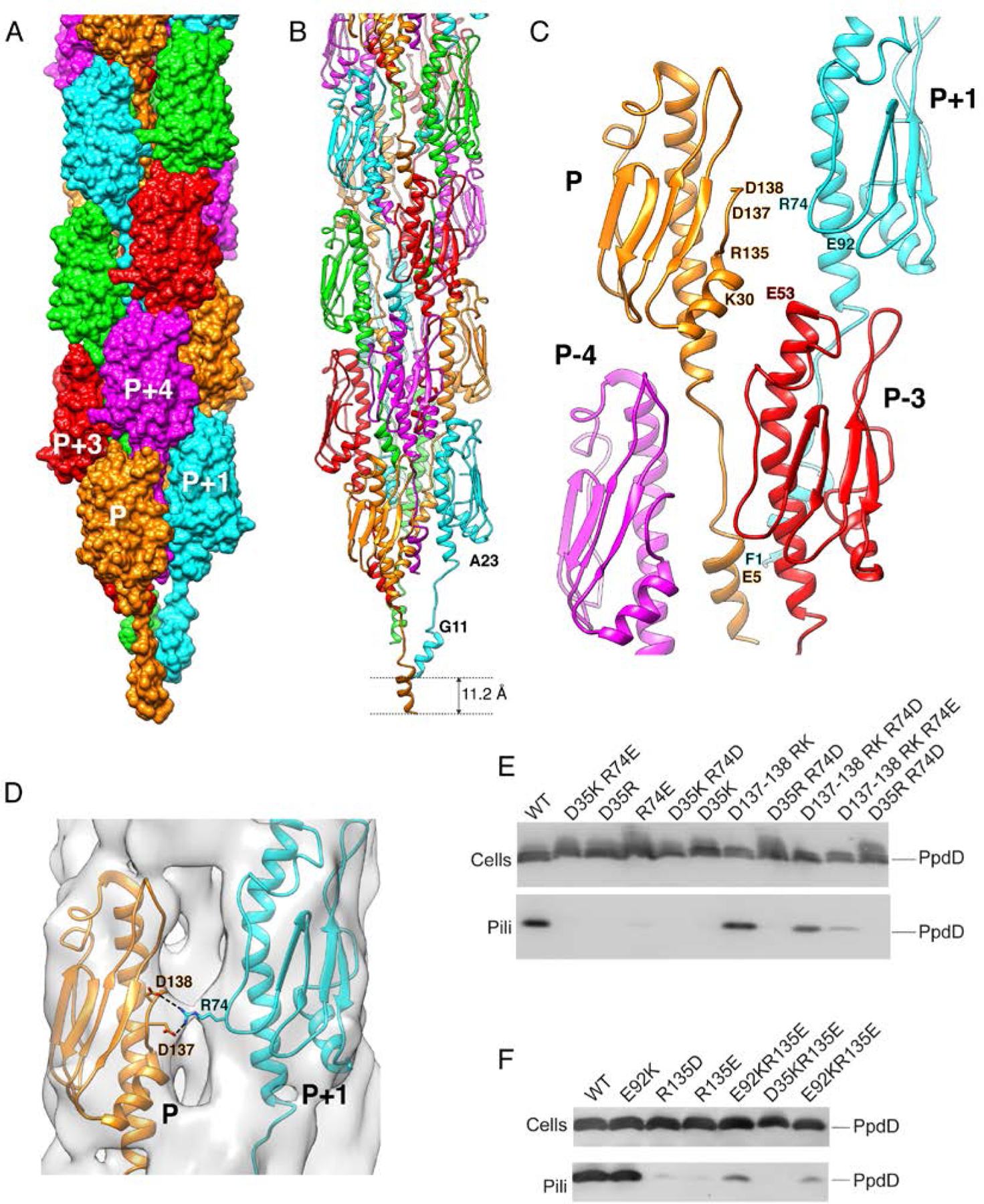
(A) Surface representation of the PpdD pilus structure with subunits P, P+1, P+2, P+3 and P+4 (along the 1-start helix) colored in orange, cyan, green, red and magenta, respectively. (B) Structure of PpdD pilus shown in ribbons where subunits are colored as in (A). The helical rise of the 1-start helix is 11.2Å. (C) Topological arrangement of neighboring subunits in the PpdD pilus structure, colored as in (A). The positions of charged residues involved in inter-subunit interactions are labeled. (D) Close-up view of the interface between protomer P and P+1 showing the side-chains of D137/D138 and R74 making potential salt-bridges in the PpdD pilus structure. The cryo-EM reconstruction is shown as surface at ~3.2σ contour level. (E-F) Piliation assay with single charge inversion PpdD variants. Cells (top) and sheared pilus fractions (Pili, bottom) corresponding to 0.05 OD600nm of bacteria were separated on SDS-PAGE and analyzed by immunoblot using anti-PpdD antibodies. PpdD residue substitutions are indicated above each lane. Migration of PpdD is indicated on the right.
