Figure 3.
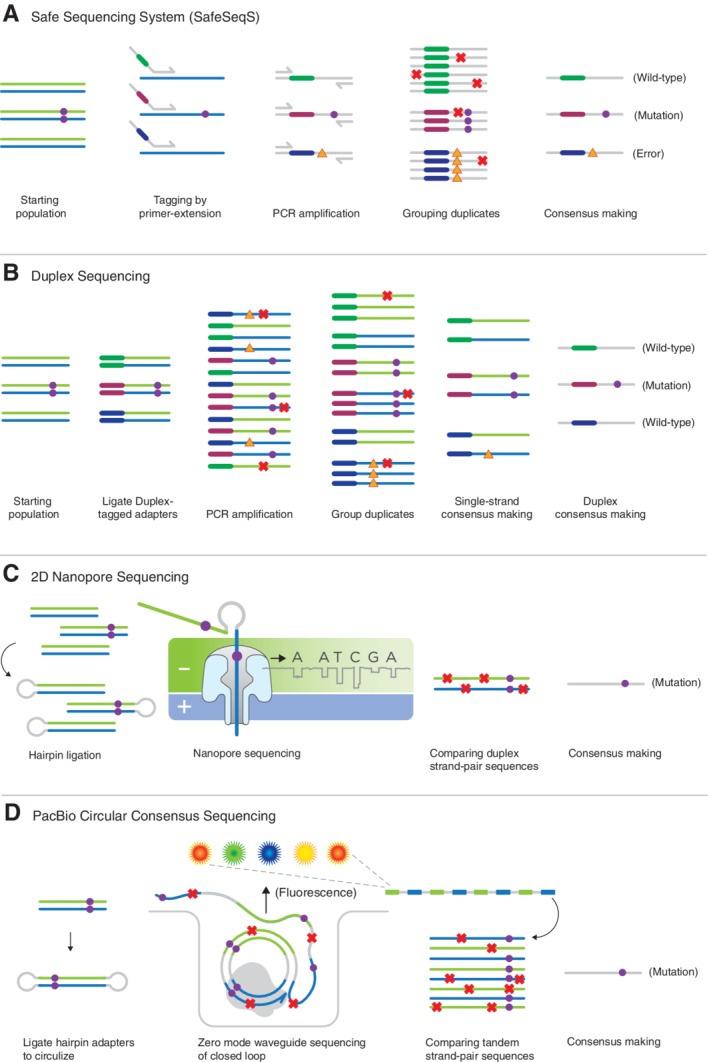
Techniques for error corrected DNA sequencing (ecNGS). The highest accuracy NGS methods rely on sequencing‐by‐consensus, whereby data from multiple sequence reads derived from an original molecule are combined to reduce the impact of sequencing or sample preparation errors in each read. (A) The SafeSeqS approach uses random molecular barcodes applied to PCR primers to uniquely tag PCR amplicons, which are then further amplified and sequenced. Variation within the sequence of reads with identical tags can be discounted as technical artifacts (X's). Some errors that occur during the first extension cycle may escape correction (triangles). (B) Duplex Sequencing relies on ligation to apply molecular barcodes to both strands of original double‐stranded molecules. These are used alone or in combination with fragmentation points to uniquely label both strands such that derivative sequence reads from each strand can be directly related back to their founder strand and compared to those from its complement. The method is significantly more accurate that single‐stranded consensus‐making methods but is more sequencing‐intensive. (C) 2D sequencing on nanopore platforms uses physical linkage of the two strands of an original duplex, which are then sequenced together without the need for amplification. The method is fast and simple, but nanopore platforms are lower accuracy and throughput than more widely used sequencing‐by‐synthesis platforms. (D) Circular Consensus Sequencing on the PacBio single‐molecule platform similarly links the two strands of an original double‐stranded with hairpins to allow multiple sequencing passes across both original strands. As with 2D, lower raw platform accuracy and throughput are drawbacks but very long reads can be obtained.
