Figure 3.
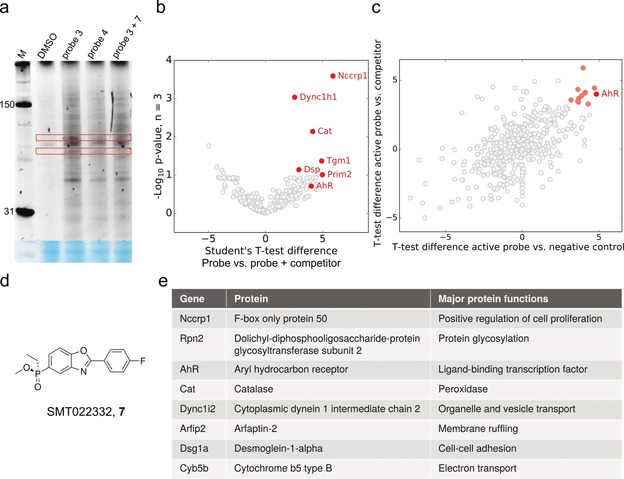
Target identification by chemical proteomics. a) In gel fluorescence of active probe 3, inactive probe 4 and active 3 with competitor 7, SMT022332 (d), labelled H2K mdx proteome. M, protein standard (in kDa); red boxes, bands that appear stronger in active probe treated samples than in the negative controls; blue stained gel represents total protein. b) Volcano plot of proteins enriched by probe 3 in the presence and absence of competitor 7, top hits in red. c) Protein enrichment by probe 3 compared to inactive probe 4 and competitor 7. Proteins strongly enriched by 3, but not the controls, in red. d) Structure of competitor 7, SMT022332. e) Top hits from (c), full list of proteins available in the supplementary information.
