Abstract
Disruption of intestinal microbial communities appears to underlie many human illnesses, but the mechanisms that promote this dysbiosis and its adverse consequences are poorly understood. In patients who received allogeneic hematopoietic cell transplantation (allo-HCT), we describe a high incidence of enterococcal expansion which was associated with graft-versus-host disease (GVHD) and mortality. We found that Enterococcus also expands in the mouse gastrointestinal tract after allo-HCT and exacerbates disease severity in gnotobiotic models. Enterococcus growth is dependent on the disaccharide lactose, and dietary lactose depletion attenuates Enterococcus outgrowth and reduces the severity of GVHD in mice. Allo-HCT patients carrying lactose-non-absorber genotypes showed compromised clearance of post-antibiotic Enterococcus domination. We report lactose as a common nutrient that drives expansion of a commensal bacterium that exacerbates an intestinal and systemic inflammatory disease.
The healthy gut is inhabited by a diverse community of mostly anaerobic bacteria, and a hallmark of microbial imbalance (dysbiosis) observed in many disease states involves the expansion of facultative anaerobic bacteria (1). Enterococci are facultative anaerobes that colonize the intestines of almost every species, from insects to mammals (2), and comprise a very small proportion (<0.1%) of the gut microbiota in healthy humans (3). However, enterococci are also pathogens; the species E. faecium and E. faecalis are a significant cause of multidrug-resistant infections in patients (4). In single-center studies, E. faecium has been observed to dominate the fecal microbiota of immunocompromised patients after allogeneic hematopoietic cell transplantation (allo-HCT), a curative-intent therapy for hematological malignancies (5–7). Moreover, fecal domination with vancomycin-resistant enterococci increases the risk of bloodstream infection in allo-HCT patients (5, 8). Patients with severe graft-vs-host disease (GVHD) after allo-HCT have poor outcomes with only ~30% long-term survival (9). Gut microbiota perturbations due to broad-spectrum antibiotics and a reduction in microbial diversity are associated with increased transplant-related mortality and lethal GVHD in humans and mice (10–14). Besides causing infections, experimental studies in gnotobiotic mice revealed that enterococci play an important role in colitis (15) by stimulating antigen-presenting cells and CD4+RORγ+ T-cell infiltration causing intestinal inflammation (16). Here, we investigated the role of enterococci in the development of acute GVHD both in allo-HCT patients and preclinical allo-HCT mouse models.
We studied the fecal microbiota of 1,325 adult allo-HCT recipients at four HCT centers: Memorial Sloan Kettering Cancer Center (MSKCC) (USA), Duke University (USA), Hokkaido University (Japan) and University Hospital Regensburg (Germany) by 16S rRNA gene sequencing. Patient characteristics are shown in table S1). We observed high abundance of enterococci early after transplantation in samples from all four transplant centers (Fig. 1B, fig. S1B). We defined Enterococcus domination as relative genus abundance ≥0.3 (≥30%) in any fecal sample, following a threshold we have used previously (5) (Supplemental Methods, fig. S2C). The incidence of domination rose comparably across centers, with up to 65% of patients showing a domination event after allo-HCT (Fig. 1A). E. faecium was the dominant species in both the MSKCC and the multicenter-validation cohort (Duke, Hokkaido, Regensburg) (Fig. 1B, fig. S1, table S2), where 40.1% of MSKCC patients (441 of 1101 patients) and 46.0% of multicenter validation patients (103 of 224 patients) met criteria for domination at any time point between day −20 and day +80 relative to the date of allo-HCT, in which cells are infused on day 0.
Fig. 1. Enterococcus domination occurs globally and increases risk of GVHD and mortality after allo-HCT.
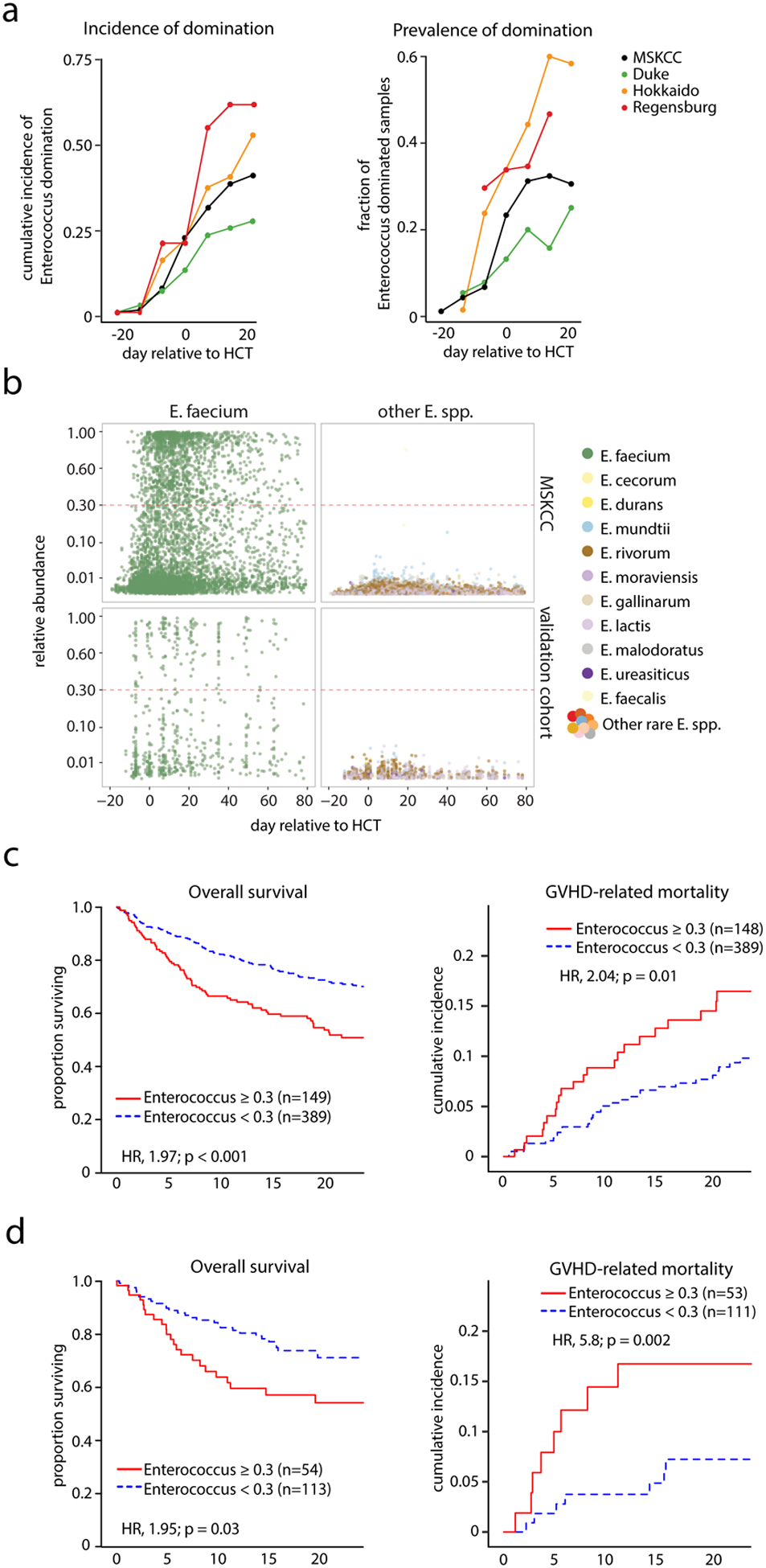
Fecal microbiota were profiled using 16S rRNA gene sequencing of 1,325 adult allo-HCT recipients. The patients attended one of four HCT centers in different countries: MSKCC (USA), Duke University (USA), Hokkaido University (Japan) and University Hospital Regensburg (Germany). (A) Left panel, cumulative incidence of patients who experienced at least one instance of genus Enterococcus domination of the gut microbiota (domination defined as a genus relative abundance of ≥0.3 (on a unitless scale from 0 to 1) over the course of allo-HCT (day −20 to +24 relative to HCT; using 7-day sliding windows) at different transplant centers; right panel, fraction of fecal specimens with enterococcal domination of the gut microbiota. Domination is defined as the relative genus abundance ≥0.3 in at least one sample between indicated days. (B) Relative abundance of different Enterococcus spp. in the microbiota of allo-HCT patients from the MSKCC and multicenter-validation cohort over the course of HCT determined by 16S rRNA gene sequencing of fecal samples; each point represents a fecal sample, color indicates the different Enterococcus spp.; the red dotted-line indicates the threshold for domination set at a relative abundance ≥0.3. (C) Overall survival and cumulative incidence of GVHD-related mortality in the T-cell replete graft recipients in the MSKCC patient cohort (see table S3), stratified into non-dominated and Enterococcus-dominated groups (domination is defined as the relative genus abundance ≥0.3 in at least one sample between day 0 and +21). (D) Overall survival and cumulative incidence of GVHD-related mortality in Enterococcus-dominated (at genus level) vs. non-dominated allo-HCT patients in the combined multicenter-validation cohort (table S3). Clinical outcomes in C and D were analyzed using the R packages survival and cmprsk. Wald p values <0.05 signify higher risks (hazard ratios, HR) of mortality among patients with Enterococcus domination as compared with those without domination.
Fecal domination by Enterococcus in the early post-transplant period (day 0 to +21) was associated with significantly reduced overall survival and increased GVHD-related mortality in both the MSKCC and multicenter-validation cohort, as well as an increased the risk of moderate-to-severe acute GVHD in the MSKCC cohort (Fig. 1C–D, fig. S2A–B, table S3). The risk of relapse/disease progression was not associated with enterococcal domination in either cohort. The association of domination by genus Enterococcus with clinical outcomes in the MSKCC remained significant in a multivariate analysis adjusted for graft source, disease, conditioning intensity, gender and age (table S4). In a subset of MSKCC patients, the vanA operon was found in 152 (37.4%) of 406 patients that had samples available for analysis, indicating the presence of vancomycin-resistant enterococci (Fig. S2E). Of note, expansions of several different taxa were detected in fecal samples in this study, but the Enterococcus genus was the most commonly observed to dominate the microbiota in all four transplant centers (fig. S3, table S5, table S6).
To further investigate these clinical observations, we examined the fecal microbiota of mice early after transplantation using well-established mouse models of allo-HCT. In a major-histocompatibility-complex (MHC)-matched minor-antigen-mismatched allo-HCT model (C57BL/6→129S1/Sv) we performed 16S rRNA gene sequencing of fecal samples and found that E. faecalis dominated the fecal microbiota at post-transplant day +8 in mice who received T-cell-replete grafts and developed lethal, acute GVHD (Fig. 2A, fig. S4A). In contrast to the patients who had prolonged antibiotic exposures, this expansion of E. faecalis was independent of antibiotic administration and dependent upon GVHD, as it was not observed in control recipients T-cell-depleted allografts in which GVHD does not develop. This post-transplant expansion of enterococci was consistently found in two additional lethal GVHD models: C57BL/6→BALB/c mice (MHC-disparate model after irradiation conditioning) (Fig. 2B), and LP/J→C57BL/6 mice (MHC-matched, minor-antigen-mismatched after busulfan/cyclophosphamide conditioning (17)) (Fig. 2C). The expansion of enterococci in murine allo-HCT recipients with GVHD was accompanied by an increase in Enterococcus colony-forming units recovered from mesenteric lymph nodes, consistent with increased bacterial translocation (Fig. 2B).
Fig. 2. Enterococcus dominates mouse gut microbiota after HCT and can exacerbate GVHD.
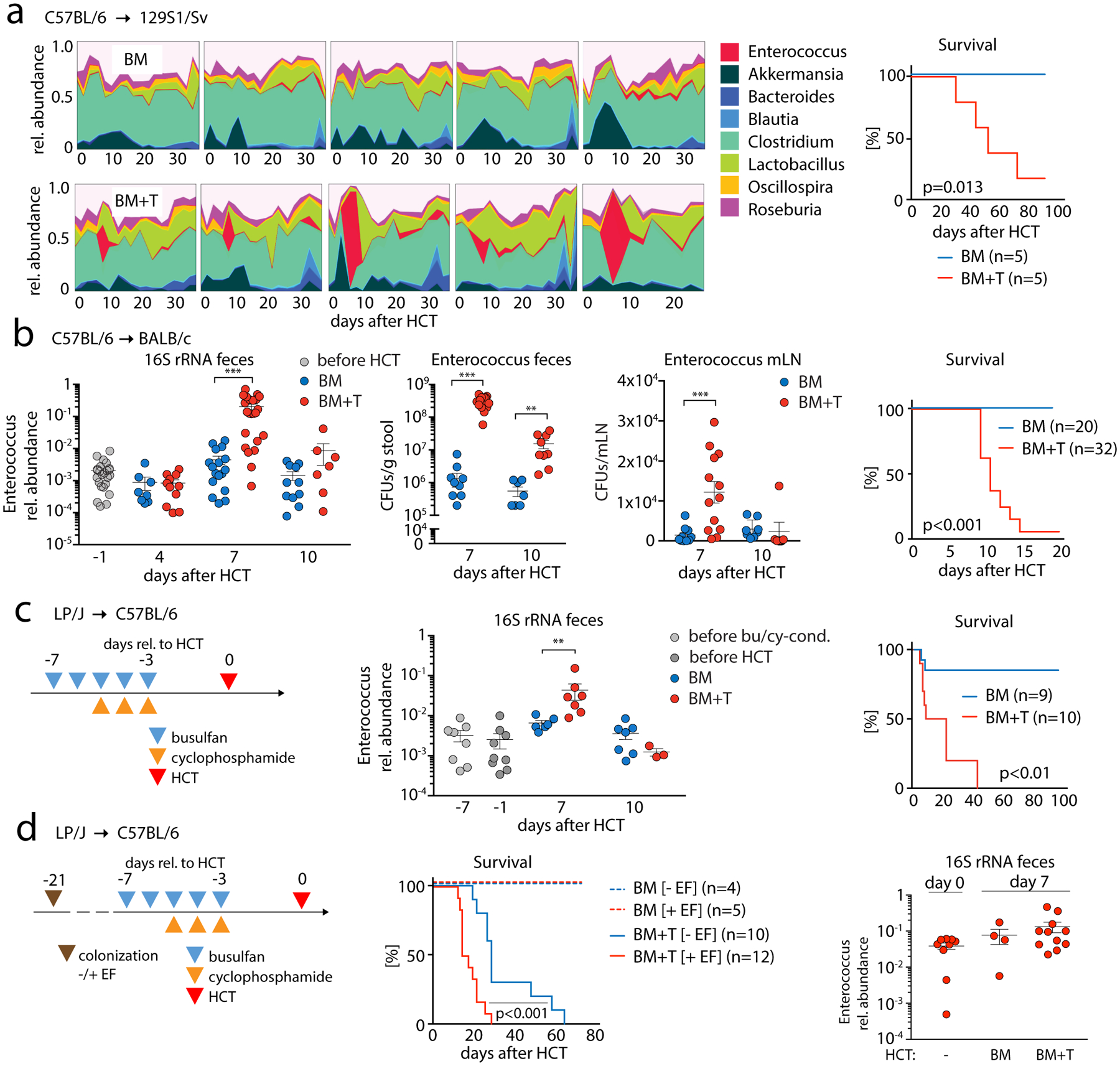
(A) High-density sampling and 16S rRNA gene sequencing of fecal microbiota from 129S1/Sv mice (1 box = 1 mouse) receiving bone marrow (BM; upper row) or T cell-replete BM (BM+T; 2×106 T cells; lower row); right panel, BM+ T cell transplanted mice develop lethal GVHD as shown by survival analysis. (B) Left panel, relative abundance of the genus Enterococcus of BALB/c host mice transplanted with C57BL/6 BM or BM+T (1×106 T cells) at different time points relative to HCT; colony forming units (CFUs) of enterococci in fecal samples and mesenteric lymph nodes (mLN). Scatter plot data show mean ± S.E.M; right panel, survival of BALB/c recipient mice after HCT (BM vs. BM + 1×106 T cells). (C) Schematic showing HCT of LP/J BM vs. BM+T (4×106 T cells) into C57BL/6 mice after chemotherapy conditioning; relative abundance of the genus Enterococcus in the feces of transplanted mice at different time points relative to HCT; right, comparison of overall survival between BM and BM+T mice. (D) Schematic showing colonization of germ-free C57BL/6 mice with a 6-strain minimal microbiota with (+EF) or without (-EF) E. faecalis OG1RF (2×107 CFUs per mouse); after 14 days, colonized mice received chemo conditioning with busulfan/cyclophosphamide and, subsequently, an HCT of LP/J BM vs. BM+T (4×106 T cells). Middle panel, comparison of overall survival; right, relative abundances of E. faecalis spiked to the minimal microbiota in the EF+ group with samples collected at the day of HCT (day 0) and 7 days later (n=4–11/group; p = 0.09, paired testing of relative abundances of enterococci of day 0 vs BM+T day 7). Scatter plot data are presented as mean ±S.E.M. **p<0.01, ***p<0.001 (independent T test for BM vs. BM+T); survival data were statistically analyzed by Mantel-Cox log-rank test.
Although we observed E. faecium domination in patients and a transient expansion of E. faecalis in GVHD mice, we hypothesized that both members of this genus might be associated with GVHD. Of note, E. faecium only recently became recognized as a major human pathogen; prior to the 1990s it was E. faecalis that caused >90% of clinical infections (18). Since E. faecalis expands in mice with GVHD and it is the major Enterococcus species in laboratory mice, we next asked whether E. faecalis contributes to GVHD. We colonized germ-free C57BL/6 mice with a community of six bacterial strains (Akkermansia muciniphila, Lactobacillus johnsonii, Blautia producta, Bacteroides sartorii, Clostridium bolteae, Parabacteroides diastonis; Supplementary Methods) (10, 19, 20) 21 days prior to allo-HCT (LP/J→gnotobiotic C57BL/6). One group of mice was co-colonized on day −21 with E. faecalis OG1RF, which remained detectable in mouse feces on days 0 and +7 (Fig. 2D right panel, fig. S4E). GVHD was exacerbated in E. faecalis-harboring mice (Fig. 2D, S4B). Serum IFNγ concentrations were significantly elevated in E. faecalis-colonized mice (fig. S4C), and we observed a significantly increased number of donor T-cells, an increase of activated and proliferating CD4+ T-cells (fig. S4D; CD4+CD25+; CD4+Ki67+) and an increased number and percentage of CD4+RORγ+ (Th17) T-cells in colon lamina propria (fig. S4D). Post-transplant administration of E. faecalis OG1RF to conventionally housed BALB/c BMT recipients also aggravated GVHD (fig. S5A). These findings indicate that E. faecalis can aggravate GVHD severity.
We next asked whether post-transplant defects of mucosal defense mechanisms facilitate enterococcal expansion. IgA-coating of intestinal bacteria can be protective in colitis and is important for maintaining mucosal integrity (21). However, we did not observe members of the genus Enterococcus to be enriched in either IgA-negative or IgA-positive fecal fractions, even though total fecal IgA was significantly reduced in allo-HCT recipients with GVHD (fig. S5B–D). Reduction in IgA by transplanting IgA-deficient BM from activation-induced cytidine deaminase (AID)-knockout mice did not further increase enterococcal expansion (fig. S5E). Intestinal antimicrobial peptides of the Reg3 family can suppress the growth of VRE (22), and are reported to play a major role in GVHD (23). Accordingly, we found that both Reg3B/G transcripts and IL-22 protein, which regulates Reg3 expression (24), were reduced in the ileum of GVHD mice (fig. S5F).
Next, we analyzed microbiota-intrinsic factors and characterized the metabolic potential of the Enterococcus-dominated fecal microbiota using shotgun metagenomic sequencing. Pre- and post-transplant fecal samples from MSKCC patients who received allo-HCT for acute myeloid leukemia were selected for sequencing on the basis of having a highly diverse pre-HCT microbiota and post-transplant E. faecium domination (by 16S rRNA gene sequencing). We focused on microbial metabolic pathways that specifically characterize domination by comparing them to the highly diverse pre-transplant microbiota from the same patients. Pathways involved in DNA synthesis and, notably, in lactose and galactose degradation were enriched in the E. faecium-dominated, post-transplant microbiota. In contrast, amino-acid synthesis and starch-degradation pathways were more prevalent in pre-transplant specimens (Fig. 3A). The lactose-and-galactose degradation pathway was also significantly enriched in the post-transplant E. faecalis-dominated samples of mice with GVHD (Fig. 3B). Comparison of whole-genome sequencing from isolates of E. faecium (from a human allo-HCT patient), and of E. faecalis (from a mouse with GVHD) revealed that genes encoding lactose and galactose metabolism accounted for ~3% of their genomes (Fig. 3C). In silico analysis of these enterococcus genomes and publicly available genomes of other members of the gnotobiotic 6-strain consortium revealed that enterococci are specifically enriched in enzymes of the tagatose-type galactose pathway for galactose to glucose degradation (25) (Supplemental Methods, fig. S6A–B). Enterococcal growth depends on lactose in vitro, as both E. faecalis and faecium strains cultured in brain-heart-infusion (BHI) broth depleted of lactose (by lactase; fig. S7A) did not grow (Fig. 3D). Growth was reinstated upon transfer to regular BHI, excluding antibacterial effects of lactase treatment (Fig. 3D). Enterococcal expansion after allo-HCT was accompanied by a loss of Clostridia spp. in the microbiota of allo-HCT patients (Figs. 3A and 3F) and of mice with GVHD (fig. S8A–C, table S7). This may be important for allo-HCT patients, as high abundances of clostridia are associated with better survival and less GVHD (12, 26). Commensal clostridia are known to produce large amounts of butyrate (27), which mitigates lethal GVHD in mice through protecting energy homeostasis of enterocytes (28). We observed that post-transplant enterococcal domination and a loss of clostridia were accompanied by a significant reduction in fecal butyrate in both allo-HCT patients and mice with GVHD (Figs. 3E, 3G; (29)). A loss of this key metabolite may contribute to the poor outcomes in Enterococcus-dominated patients and mice.
Fig. 3. Metagenomic and metabolomic analyses of Enterococcus-dominated fecal specimens in human HCT patients and mice.
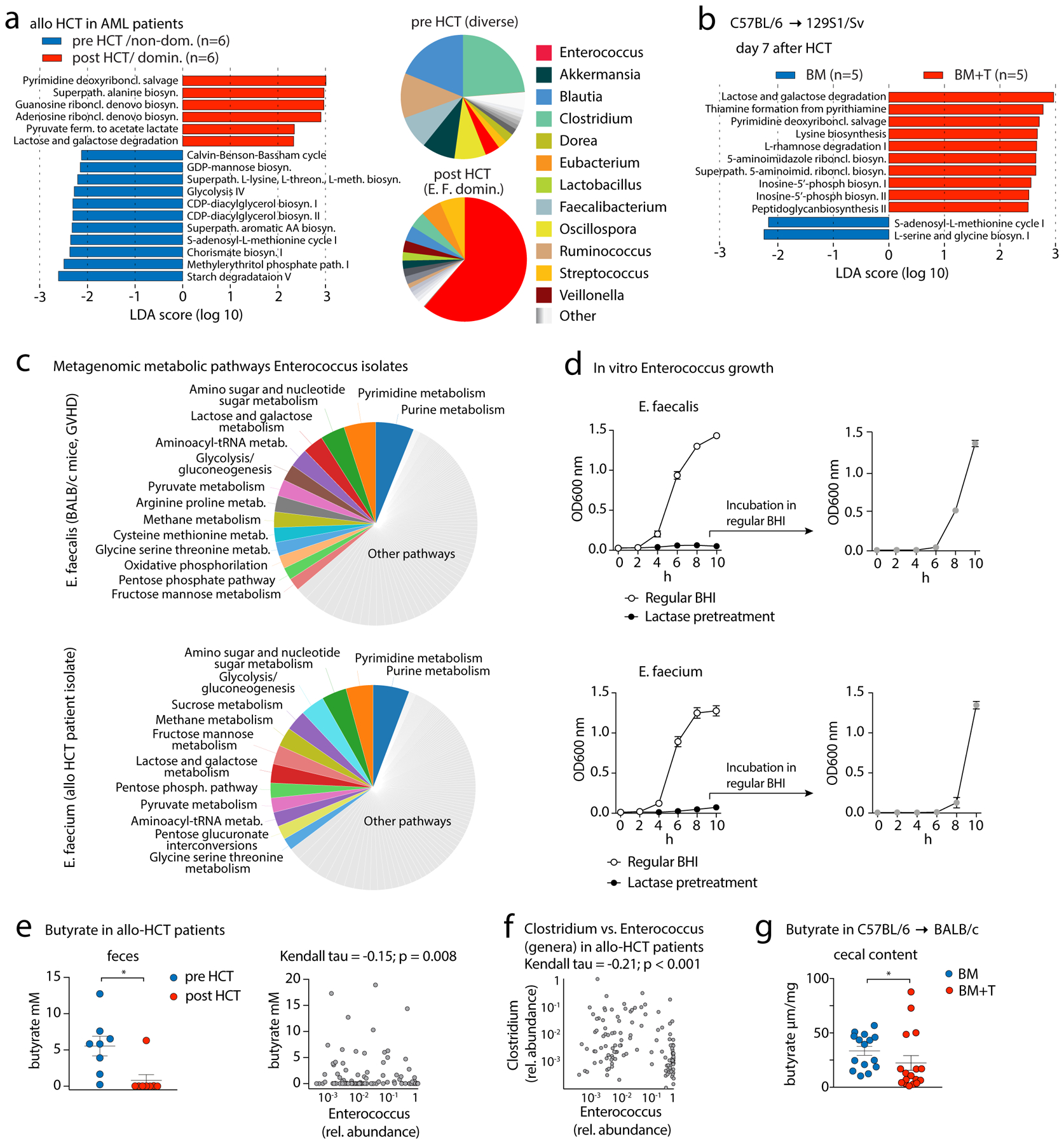
(A) Left, differential abundances of shotgun-sequenced and HUMAnN2-annotated bacterial metabolic pathways between paired pre- and post-HCT fecal samples from MSKCC patients who received allo-HCT for acute myeloid leukemia (AML) analyzed by linear discriminant analysis (LDA) coupled with effect size measurements (LEfSe). (definitions: pre: day −8 to −1; post: day 3–25 after allo-HCT). Right panel, pie chart showing mean relative abundances of bacterial genera (analyzed by MetaPhlAn2) found in patient fecal samples pre- and post-HCT; data are aggregated across all patients. (B) LEfSe analysis of bacterial metabolic pathway abundances in HCT day +7 fecal samples of 129S1/SV mice transplanted with C57BL/6 BM vs BM+T (2×106 T cells) (see Fig. 2A). (C) Left panel, pie chart with metabolic pathway abundances determined by whole genome sequencing of E. faecalis (isolated from feces of a BALB/c GVHD mouse, day +7 after HCT) and E. faecium (human allo-HCT fecal isolate; only pathways with an abundance ≥ 2% are shown). (D) Left panel, in vitro growth of E. faecalis (mouse GVHD isolate; upper panel) and E. faecium (human allo-HCT isolate; lower panel) in non-treated BHI broth or in BHI broth pretreated with lactase. Right panel, E. faecalis or E. faecium incubated in lactase-pretreated BHI were put into regular BHI broth after 8h to assess growth dynamics in regular BHI (grey symbol); 4 experiments combined, values represent mean ±S.E.M. (E) Left panel, fecal butyrate concentrations (mean ±S.E.M.) from pre- and post-transplant fecal samples from AML patients from MSKCC who received allo-HCT and were selected on a highly diverse pre-HCT microbiota and a post-transplant E. faecium domination (by 16S rRNA gene sequencing; 6 (out of 8) patients are presented in Fig. 3A; right panel), correlation of butyrate concentrations with relative abundances of the genus Enterococcus (n=139 patients; 8 patients from Fig. 3E in the left panel), and 131 allo-HCT patients from a dataset published by Haak et al. (29); statistical analysis was performed using Kendall’s tau rank correlation coefficient. (F) Stool samples were collected at the time of engraftment (~24 days after allo-HCT). Data show Kendall’s tau rank correlation of relative abundances of the genera Clostridium and Enterococcus from the data-set of Haak et al. (G) Butyrate concentration (mean ±S.E.M.) in cecal contents of BALB/c mice transplanted with C57BL/6 BM or BM+T (1×106 T cells) at day 7 after HCT. Statistical analysis: *p<0.05 (paired T-test (E) or independent T-test for (F)).
Given that the optimal growth of enterococci depends on lactose availability in vitro, we asked whether enterococcal expansion can be mitigated by feeding mice lactose-free chow (fig. S7B; table S8). In the C57BL/6→BALB/c model, the absence of dietary lactose significantly reduced post-transplant Enterococcus bloom and mitigated GVHD (Fig. 4A, fig. S9B). Flow cytometric analysis of donor T-cells on day +14 revealed a reduction in the percentage of activated and proliferating CD4+ T-cells (CD4+CD69+; CD4+Ki67+) as well as a reduction in the percentage of CD4+Tbet+ (Th1) T-cells (fig. S9). The effect of a lactose-free diet on enterococcal outgrowth and GVHD was replicated in the LP/J→C57BL/6 mouse model (Fig. 4B; table S9 for changes in non-enterococcal taxa). Intestinal mucosal damage by irradiation or allo-reactive T-cells may affect the expression of lactase, the enzyme found on small-intestine enterocytes to facilitate lactose absorption through disaccharide cleavage. Duodenal lactase transcript abundance progressively declined in BM+T recipients over the course of transplantation (fig. S9c), which may render mice to a “lactose-intolerant”-like state, allowing non-digested lactose to reach the lower intestinal tract and serve as a carbon source for bacteria.
Fig. 4. Lactose-free diet reduces experimental GVHD and lactase genotypes associated with microbiota dynamics after allo-HCT in humans.
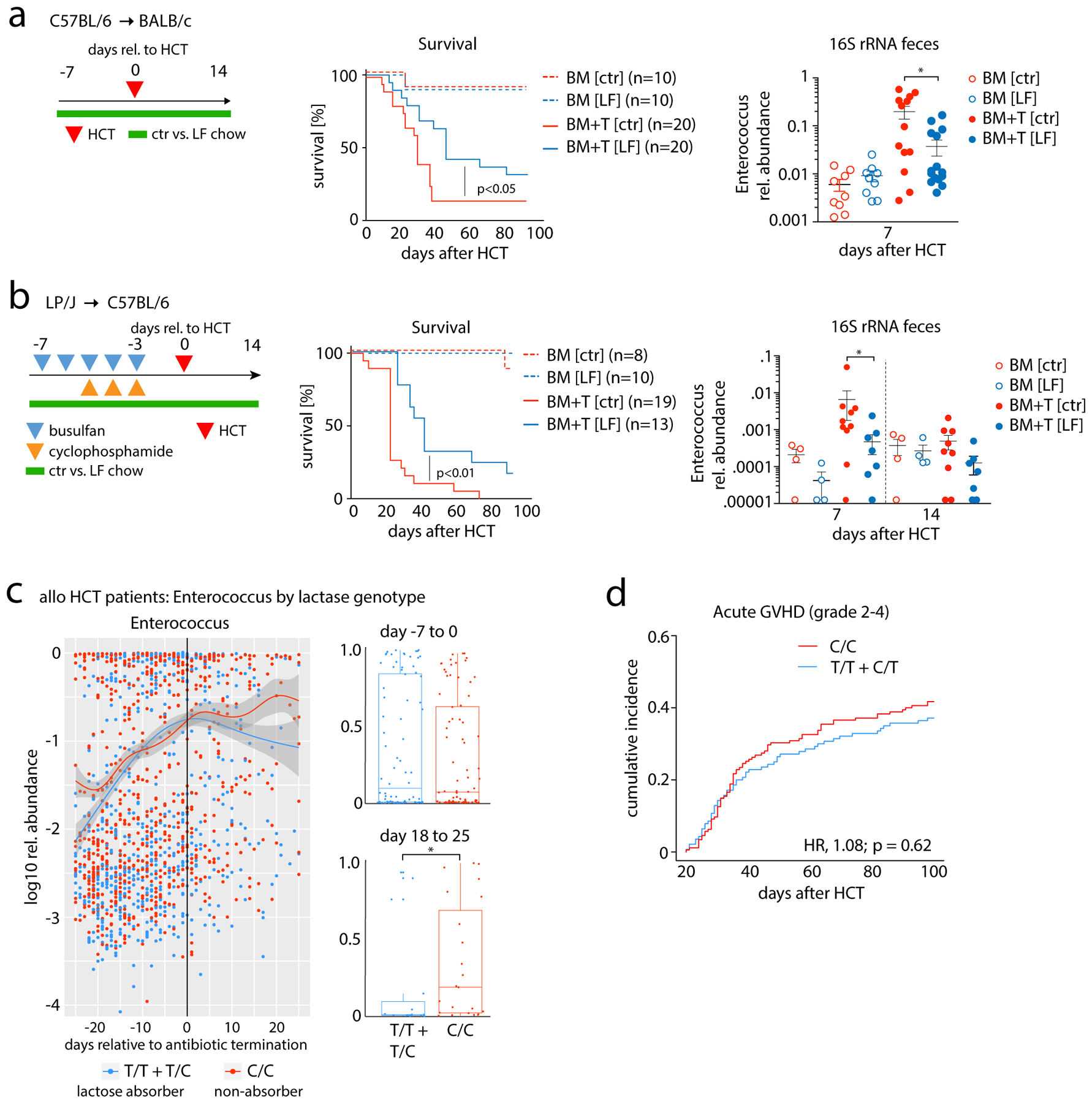
(A) Schematic showing that BALB/c recipient mice received C57BL/6 BM or BM + T (5×105 T cells) and were fed control chow (ctr) vs. lactose-free chow (LF) from day −7 to 14 relative to transplant; comparison of survival between BM and BM+T mice (middle panel) and relative abundance of the genus Enterococcus in mouse feces (right panel). Scatter plot data presented as mean ±S.E.M; *p<0.05 (independent T test). (B) Schematic showing HCT of LP/J BM vs. BM+T (4×106 T cells) into C57BL/6 mice after chemotherapy conditioning; comparison of survival between BM and BM+T mice (middle panel) and relative abundance of the genus Enterococcus at different time points relative to HCT (right panel). Scatter plot data presented as mean ±S.E.M; *p<0.05 (independent T test) (C) Left, relative abundance (log10) of Enterococcus (genus) by days relative to the day of antibiotic cessation (broad-spectrum antibiotics for neutropenic fever: either piperacillin-tazobactam i.v., imipenem-cilastatin i.v., or meropenem iv); box plot-inserts display the median relative abundances of the genus Enterococcus of time binned in the indicated day ranges relative to antibiotic cessation; whiskers represent maximum and minimum. Statistical analysis of box plot data: *p<0.05, ***p<0.001 (Wilcoxon rank test). (D) Cumulative incidence of acute GVHD grade 2–4 in rs4988235 SNP-genotyped MSKCC patients (T cell-depleted grafts excluded; graft source: BM/PBSC unmodified = 213 patients; cord blood = 102 patients; C/C = 175, T/C+T/T = 140). The cumulative incidence of Grade 2–4 acute GVHD was compared between genotype groups using the R package cmprsk. HR, hazard ratio.
Next, we asked whether enterococci expansion is associated with lactose tolerance in human allo-HCT patients by genotyping 602 patients from the MSKCC cohort with available pre-transplant germline DNA samples for the gene polymorphism rs4988235 (−13910*T). This SNP regulates lactase expression and predicts lactose absorption/tolerance (C/T- or T/T-alleles) and malabsorption (C/C-alleles) in the upper gut (30). While abundance of the genus Enterococcus increased comparably during exposure to broad-spectrum antibiotics in both lactose absorbers and malabsorbers, enterococcal domination was significantly prolonged in malabsorbers after cessation of antibiotics (Fig. 4D, fig. S10). This finding suggests that the maintenance of enterococcal domination and microbiota recovery after broad-spectrum antibiotic exposure is significantly modulated by the luminal availability of lactose as a growth substrate.
In conclusion, fecal domination by Enterococcus spp. is a significant risk factor for the development of acute GVHD, for increased overall and GVHD-related mortality after allo-HCT. Our findings significantly extend previous reports from smaller single-center analyses that post-transplant VRE bacteremia and fecal domination are associated with worse outcomes after allo-HCT (7, 8, 31). In gnotobiotic mouse models, enterococci exacerbate GVHD, consistent with previous reports of aggravated colitis in models of inflammatory bowel disease (15) or systemic autoimmune responses (32). We previously identified Blautia abundance (a genus within class Clostridia) as a predictor of protection from lethal GVHD (12), while here we describe that Enterococcus domination is a risk factor for GVHD. These two findings are interesting in light of our recent observation that a Blautia producta strain can inhibit VRE growth via the production of a lantibiotic protein (33). We identified a microbiota-intrinsic mechanism that is dependent on lactose utilization and favors the expansion of enterococci. This process may be triggered through a loss of lactase produced by enterocytes damaged by conditioning or allo-reactive T-cells. We validated this concept experimentally by showing that depletion of lactose in vitro and in vivo inhibited enterococcal expansion and mitigated GVHD, and clinically by showing that patients harboring a lactose-malabsorption allele experienced prolonged Enterococcus domination after antibiotic exposure. These observations in mice and allo-HCT patients provide proof-of-concept for a novel, non-antibiotic-based therapeutic strategy such as a lactose-free diet to attenuate the outgrowth of pathobionts like enterococci and possibly improving clinical outcomes by modulating dietary sources of nutrients for pathogenic bacteria.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank G.M. Dunny and J. Willett, Department of Microbiology, Univ. Minnesota, as well as R. Zbasnik and V. Schlegel, Department of Food Science, Univ. Nebraska, for helpful discussion and providing materials for revision of the manuscript.
Funding: This work was supported by a German Research Foundation (DFG) scholarship to C.S.T., a Young-Investigator-Award by American Society of Bone Marrow Transplantation to C.S.T.; partially supported by the DFG research consortium TR221 “GvH/GvL” (project B13) to E.H.; by NCI awards, R01-CA228358 (M.V.D.B..), R01-CA228308 (M.V.D.B.), MSKCC Cancer Center Core Grant P30 CA008748, and Project 4 of P01-CA023766 (M.V.D.B.); NHLBI award R01-HL125571 (M.V.D.B.) and R01-HL123340 (M.V.D.B.); NIA National Institute of Aging award Project 2 of P01-AG052359 (M.V.D.B.); NIAID award U01 AI124275 (M.V.D.B.); R01 AI032135 (E.G.P.); AI095706 (E.G.P.); U01 AI124275 (E.G.P. and J.B.X.); Tri-Institutional Stem Cell Initiative award 2016-013 (M.V.D.B.); The Lymphoma Foundation (M.V.D.B. and N.K.); The Susan and Peter Solomon Divisional Genomics Program (M.V.D.B.); and the Parker Institute for Cancer Immunotherapy at Memorial Sloan Kettering Cancer Center (M.V.D.B. and J.U.P.); the Sawiris Foundation (J.U.P.); the Society of Memorial Sloan Kettering Cancer Center (J.U.P.); MSKCC Cancer Systems Immunology Pilot Grant (J.U.P.); Empire Clinical Research Investigator Program (J.U.P.); Seres Therapeutics (M.V.D.B., J.U.P., J.B.S., A.L.C.G., A.G.C., and A.E.S., A.D.S.); Japan Society for the Promotion of Science KAKENHI (17H04206 to T.T., 17K09945 to D.H.); The Center of Innovation Program from Japan Science and Technology Agency (T.T.); Mochida Memorial Foundation for Medical and Pharmaceutical Research (D.H.); R56 AI137269-01 (J.B.X.); Conquer Cancer Foundation Young Investigator Award/Gilead Sciences (N.K.); NIH KL2 TR001115-03 (NCATS CTSA to A.D.S.), NIA 2P30AG028716-11 (Claude D. Pepper Older Americans Independence Center to A.D.S.); NCI R01CA203950-01 (to N.J.C., A.D.S., L.B., M.L, and A.B.); NIH 1R01HL124112-01A (A.D.S. and R.R.J.); NCI R01 CA203950-01 (A.D.S. and N.J.C.).
Footnotes
Competing interests: M.V.D.B. is on the advisory board of and has financial holdings in Seres Therapeutics Inc., serves as DKMS chairman, has received speaker honoraria from Merck and Acute Leukemia Forum, holds patents that receive royalties from Seres Therapeutics Inc., has received honorarium and research support (1 January 2017 to present) from Seres Therapeutic Inc., and IP licensing with Seres Therapeutics Inc. and Juno. J.U.P. reports research funding, intellectual property fees, and travel reimbursement from Seres Therapeutics and consulting fees from Davolterra. E.G.P. has received speaker honoraria from Bristol-Myer Squibb, Celgene, Seres Therapeutics, MedImmune, Novartis, and Ferring Pharmaceuticals; is an inventor on patent application no. WPO2015179437A1, entitled ‘Methods and compositions for reducing Clostridium difficile infection’ and no. WPO2017091753A1, entitled ‘Methods and compositions for reducing vancomycin-resistant Enterococci infection or colonization’; and holds patents that receive royalties from Seres Therapeutics Inc.. Other authors have no competing interests.
Data and materials availability:
All data are available in the manuscript or the supplementary materials. Sequencing data are deposited into SRA under Bioproject number PRJNA545312.
REFERENCES
- 1.Litvak Y, Byndloss MX, Baumler AJ, Colonocyte metabolism shapes the gut microbiota. Science (New York, N.Y.) 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilmore MS, Lebreton F, van Schaik W, Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr Opin Microbiol 16, 10–16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schloissnig S et al. , Genomic variation landscape of the human gut microbiome. Nature 493, 45–50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lebreton F et al. , Tracing the Enterococci from Paleozoic Origins to the Hospital. Cell 169, 849–861.e813 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taur Y et al. , Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 55, 905–914 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ubeda C et al. , Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 120, 4332–4341 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holler E et al. , Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transplant 20, 640–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ford CD et al. , Vancomycin-Resistant Enterococcus Colonization and Bacteremia and Hematopoietic Stem Cell Transplantation Outcomes. Biol Blood Marrow Transplant 23, 340–346 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Khoury HJ et al. , Improved survival after acute graft. Haematologica 102, 958–966 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shono Y et al. , Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci Transl Med 8, 339ra371 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taur Y et al. , The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 124, 1174–1182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenq RR et al. , Intestinal Blautia Is Associated with Reduced Death from Graft-versus-Host Disease. Biol Blood Marrow Transplant 21, 1373–1383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peled JU et al. , Microbiota as a predictor of mortality in allogeneic HCT submitted. [DOI] [PMC free article] [PubMed]
- 14.Taur Y et al. , Reconstitution of the gut microbiota of antibiotic-treated patients by autologous fecal microbiota transplant. Sci Transl Med 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steck N et al. , Enterococcus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation. Gastroenterology 141, 959–971 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Geva-Zatorsky N et al. , Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 168, 928–943.e911 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riesner K, Kalupa M, Shi Y, Elezkurtaj S, Penack O, A preclinical acute GVHD mouse model based on chemotherapy conditioning and MHC-matched transplantation. Bone Marrow Transplant 51, 410–417 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Dubin K, Pamer EG, Enterococci and Their Interactions with the Intestinal Microbiome. Microbiology spectrum 5, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenq RR et al. , Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. The Journal of experimental medicine 209, 903–911 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caballero S et al. , Cooperating Commensals Restore Colonization Resistance to Vancomycin-Resistant Enterococcus faecium. Cell Host Microbe 21, 592–602.e594 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palm NW et al. , Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brandl K et al. , Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455, 804–807 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao D et al. , Survival signal REG3α prevents crypt apoptosis to control acute gastrointestinal graft-versus-host disease. J Clin Invest 128, 4970–4979 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindemans CA et al. , Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature 528, 560–564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schleifer KH, Hartinger A, Götz F, Occurence of D-tagatose-6-phosphate pathway of D-galactose metabolism among staphylococci. FEMS Microbiology Letters 3, 9–11 (1978). [Google Scholar]
- 26.Weber D et al. , Microbiota Disruption Induced by Early Use of Broad-Spectrum Antibiotics Is an Independent Risk Factor of Outcome after Allogeneic Stem Cell Transplantation. Biol Blood Marrow Transplant 23, 845–852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gill PA, van Zelm MC, Muir JG, Gibson PR, Review article: short chain fatty acids as potential therapeutic agents in human gastrointestinal and inflammatory disorders. Aliment Pharmacol Ther 48, 15–34 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Mathewson ND et al. , Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat Immunol 17, 505–513 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haak BW et al. , Impact of gut colonization with butyrate-producing microbiota on respiratory viral infection following allo-HCT. Blood 131, 2978–2986 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ingram CJ, Mulcare CA, Itan Y, Thomas MG, Swallow DM, Lactose digestion and the evolutionary genetics of lactase persistence. Hum Genet 124, 579–591 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Vydra J et al. , Enterococcal bacteremia is associated with increased risk of mortality in recipients of allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 55, 764–770 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manfredo Vieira S et al. , Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science (New York, N.Y.) 359, 1156–1161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SG et al. , Microbiota-derived lantibiotic restores resistance against vancomycin-resistant Enterococcus. Nature 572, 665–669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


