Figure 1.
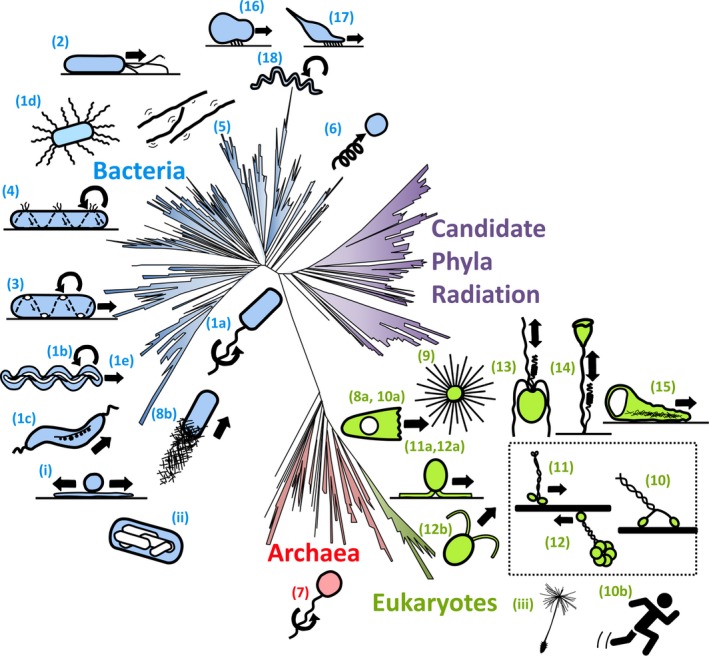
Various types of motility systems. Cartoons of those systems are listed according to the order in the text and roughly assigned to the relative positions in Tree of Life (Hug et al. 2016; Castelle & Banfield 2018). (1a) bacterial flagellar swimming, (1b) spirochetes flagellar swimming, (1c) magnetotactic bacterial flagellar swimming, (1d) bacterial flagellar swarming, (1e) Leptospira crawling motility, (2) bacterial pili motility, (3) Myxococcus xanthus adventurous (A) motility, (4) Bacteroidetes gliding, (5) Chloroflexus aggregans surface motility, (6) Synechococcus nonflagellar swimming, (7) archaella swimming, (8a) amoeba motility based on actin polymerization, (9) heliozoa motility based on microtubule depolymerization, (10) myosin sliding, (11) kinesin sliding, (12) dynein sliding, (10a) amoeba motility driven by contraction of cortical actin–myosin. (10b) animal muscle contraction, (11a, 12a) flagellar surface motility (FSM), (12b) flagellar swimming, (13) haptonemal contraction, (14) spasmoneme contraction, (15) amoeboid motility of nematode sperm, (8b) actin‐based comet tail bacterial motility, (16) Mycoplasma mobile gliding, (17) Mycoplasma pneumoniae gliding, (18) Spiroplasma swimming, (i) bacterial sliding, (ii) gas vesicle, (iii) dandelion seed. Refer to Table 1 for more details. The three eukaryotic conventional motor proteins are shown in the dotted box
