Abstract
Obesity is a multifactorial disease caused by complex interactions between genes and dietary factors. Salt-rich diet is related to the development and progression of several chronic diseases including obesity. However, the molecular basis of how salt sensitivity genes (SSG) contribute to adiposity in obesity patients remains unexplored. In this study, we used the microarray expression data of visceral adipose tissue samples and constructed a complex protein-interaction network of salt sensitivity genes and their co-expressed genes to trace the molecular pathways connected to obesity. The Salt Sensitivity Protein Interaction Network (SSPIN) of 2691 differentially expressed genes and their 15474 interactions has shown that adipose tissues are enriched with the expression of 23 SSGs, 16 hubs and 84 bottlenecks (p = 2.52 x 10–16) involved in diverse molecular pathways connected to adiposity. Fifteen of these 23 SSGs along with 8 other SSGs showed a co-expression with enriched obesity-related genes (r ≥ 0.8). These SSGs and their co-expression partners are involved in diverse metabolic pathways including adipogenesis, adipocytokine signaling pathway, renin-angiotensin system, etc. This study concludes that SSGs could act as molecular signatures for tracing the basis of adipogenesis among obese patients. Integrated network centered methods may accelerate the identification of new molecular targets from the complex obesity genomics data.
Introduction
Obesity, an excessive body fat accumulation in individuals acts as a major risk factor for the development of diverse chronic diseases like impaired insulin metabolism, glycemic abnormalities, hypertension and cardiovascular diseases in future. Obesity, owing to its complex multifactorial disease nature is not only challenging the molecular scientists to decode its molecular basis but also the clinicians who are involved in treating, preventing and disease management. Approximately 30% of the world population is either overweight or obese [1]. So far, the specific molecular and cellular mechanisms through which environmental factors increase the risk of developing obesity in genetically susceptible individuals still remains to be a mystery. The chronic low inflammation in different tissues is one of the characteristic features of obesity [2]. Particularly, chronic inflammatory reactions which takes place in adipose tissues contribute to the obesity associated insulin insensitivity. Adipose tissue plays an important role in the development of metabolic diseases due to dysregulated discharge of adipocytokines from adipocytes in visceral fat of obese individuals. This will subsequently induce insulin resistance condition in muscles and liver. The faulty insulin sensitivity of adipose tissues, connects the obesity with other chronic diseases like diabetes, hyperlipidemia, arthritis, hypertension, cardiovascular disease, ischemic stroke, hyperglycemia and different types of cancer [3] [4].
The importance of excess salt intake in the pathogenesis of metabolic diseases is widely recognized. Salt sensitivity is a physiological trait, in which the changes in salt intake parallel the changes in blood pressure [5]. The gene expression status of salt sensitivity genes (SSGs) in adipose tissues is not yet well explored. In the present study, we focused on SSGs expressed in adipose tissues to figure their influential role in the pathogenesis of obesity. We considered genes from renin-angiotensin system pathway which maintains the homeostasis of salt and body fluids, and regulate the blood pressure [6]. In addition, expression of renin-angiotensin system in adipose tissue is involved in the regulation of triglyceride accumulation, adipocyte formation, glucose metabolism, lipolysis, and the initiation of the adverse metabolic consequences of obesity [7], [8]. Therefore, in order to identify the candidate genes from SSGs and their molecular signature networks connected to the pathogenesis of obesity, the gene expression datasets collected from visceral adipose tissues were analyzed by knowledge based systemic investigations and statistical methods. We used different statistical parameters like graph theory to pick up biomarkers from the gene expression data. We also used gene-gene correlation, which relies on the fact that disease candidate genes showing a similar expression pattern are more likely to interact with one another for their biological functioning[9]. Our network biology integrated investigation will offer novel association with potential biological comprehensions and supports future translational assessment on SSGs and obesity.
Materials and methods
Gene expression dataset
The microarray generated gene expression dataset with the reference ID of GSE88837 was collected from GEO (Gene Expression Omnibus) database [10]. This gene expression data is generated on Affymetrix microarray platform using the total RNA extracted from human visceral adipose tissue of 16 overweight woman adolescent samples (BMI > 25) and 14 lean adolescent women (BMI < 25). Complete information about the individuals and testing methods, can be found in S1 Table.
Normalization of gene expression data
Gene expression data analysis of the samples were implemented by means of R packages [11] [12]. For the standardization and noise reduction in the probe data, CEL files were incorporated into R package, Affy, and the unprocessed signal intensity values of each gene expression probe sets were standardized with help of a statistical algorithm called as RMA (Robust Multiarray Average). This RMA algorithm performs the normalization of raw intensity data by generating a matrix of gene expression data whose background is corrected and log2 conversion, and then quantile normalization was performed [12]. The standardized samples were then quantitatively categorized as normal (control) and obese (disease) sets. The statistical difference between differentially expressed genes (DEGs) was computed using unpaired t-test measure among healthy and obese samples [13]. For examine the statistical differences in DEGs, the false discovery rate of Benjamini and Hochberg with a p value of 0.05 was conducted[14].
Building network of proteinprotein interaction
Bisogenet, a cytoscape plugin, was used to derive associations between the DEGs obtained from the profiles of expression. Bisogenet finds significant gene interactions from high-performance experiments and deposited literature data in DIP (Database of Interacting Proteins), BIND (Biomolecular Interaction Network Database), BioGRID (Biological General Repository for Interaction Datasets), MINT (The Molecular Interaction database), HPRD (Human Protein Reference Database), and IntAct databases [13, 15–19].
Construction of subnetwork
The complex interactome Protein Interaction Network (PIN) was rescaled to a significant subnetwork of Salt Sensitivity Protein Interaction Network (SSPIN) by following admitted notions in the network biology. From the Protein Interaction Network, we extracted genes that belong to (a) hubs based on degree centrality (DC), (b) betweenness centrality (BC) based bottlenecks (c) salt sensitivity genes. The PIN created from Bisogenet was optimized and imported to Cytoscape 3.2.1 in order to represent and measure the different parameters like DC and BC connected to network centrality of each individual protein in the biological network [20]. The Network Analyzer [21] Cytoscape plugin was deployed to monitor the network's local and global centrality parameters [14, 22–24].
Selection of hub proteins
DC of a gene is the number of partners that are connected to that specific gene. Genes which shows higher DC in any given biological network will possess many interacting partners[25]. In PIN, genes having higher DC corresponds to essential genes. For identifying the hubs, we followed the hub classification approach, which was previously described by Rakshit et al., [26]. The cut-off scores used for DC, while selecting the hub protein is described as:
| (Formula 1) |
where Avg is the average DC of significantly expressed genes in the PIN and SD denotes the standard deviation values [26].
Identification of bottlenecks
The higher DC is in correspondence to biologically essential genes, but DC is unable to quantify significance of any gene in a network [27]. Based on the theory of the protein's local property, DC does not assess the global value of the protein in the network. There could be several other key indicators that show the importance of a protein in the network based on its global significance. A global BC measure was therefore implemented to determine the characteristics of any query gene at the entire interactome level [28]. BC is measured by applying following formula:
| (Formula 2) |
where ‘s’ and ‘t’ are the network nodes, other than ‘n’ and σst(n) is the number of shortest paths from s to t that ‘n’ lies upon [29]. The significantly expressed genes falling in top 25% are regarded under bottleneck category using the node betweenness distribution.
Salt sensitivity genes
The genes involved in the pathway of renin angiotensin aldosterone system were collected as they serve as chief component in the regulation of salt and water balance of the body [30]. We also collected salt sensitivity genes from a detailed literature survey [31] [5] [32] [33]. In total, we obtained 47 SSG as represented in S2 Table.
Mapping of weighted gene-gene correlations
The map detailing gene-gene correlations was created on the basis of the algorithm known as the Pearson correlation across the entire gene set in the SSPIN. The “r” value indicating the correlation between gene pairs in the expression data was generated with help of Pearson’s correlation coefficient (PCC) method. The formula used for calculating PCC for gene pairs is described in below given Formula 3.
| (Formula 3) |
where and indicates average of the expression values of two genes in the samples, respectively.
Functional enrichment analysis
Functional enrichment analysis validates the physiological importance of the genes involved in a biological process and helps to reveal unintended gene activity. ToppGene Suite was employed to perform functional enrichment of the filtered genes [34].
Results
Microarray gene expression profile analysis
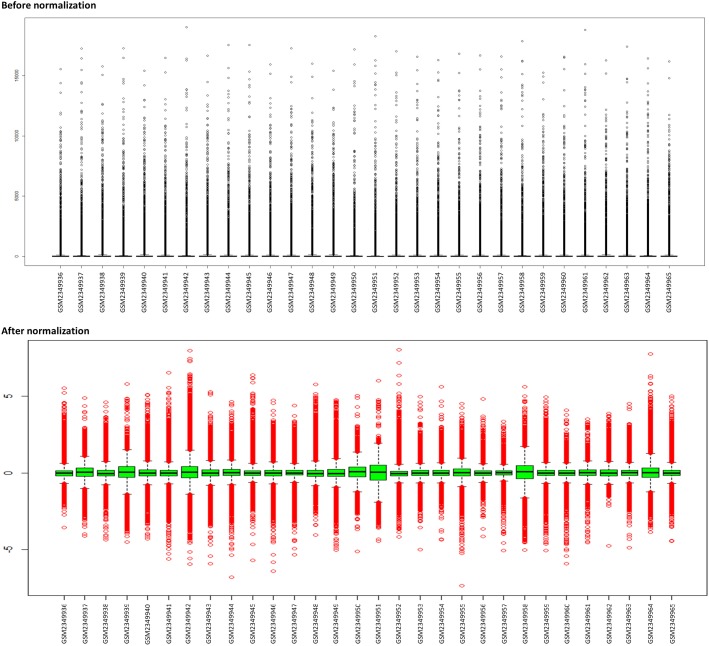
We obtained 2691 significant genes from the analysis of raw gene expression signals using RMA with statistical significance of p value ≤ 0.05. The intensity values of genes in the expression profiles, before and after normalization, are depicted as box plots which represents standardized form of representing the data distribution in Fig 1.
Fig 1. Pre and post-normalization of microarray gene expression data.
Samples are represented on horizontal axis and the gene expression values on vertical axis.
Constructed Protein Interaction Network
Overall 2691 differentially expressed genes generated from the microarray expression profile were inputted in Bisogenet, a plugin in Cytoscape, to create PIN by extracting all potential connectivity between the genes. The created PIN comprised of outliers like replicated edges and self-loops. The PIN is transformed to a stable network by eliminating self-loops and replicated edges which is then used to calculate the standardized graph centrality parameters for each single gene. The plugin created a complex PIN, covered of 2691 nodes and 15474 edges with edge-node ratio of 5.75 on an average. Next, the plugin NetworkAnalyzer, calculated the degree centrality betweenness centrality parameters of the network which are considered as local and global graph parameters respectively [21]. Table 1 provides a description of the top 10 significant genes dependent on the highest degree centrality along with general parameters of centrality.
Table 1. List of 10 significant genes obtained from network analysis based on graph theory.
| Gene | Name | BC# | DC$ |
|---|---|---|---|
| PHF8 | PHD Finger Protein 8 | 0.260 | 882 |
| EGR1 | Early Growth Response 1 | 0.091 | 504 |
| JUND | Jund Proto-Oncogene, Ap-1 Transcription Factor Subunit | 0.089 | 462 |
| FOS | Fos Proto-Oncogene, Ap-1 Transcription Factor Subunit | 0.076 | 438 |
| CHD2 | Chromodomain Helicase Dna Binding Protein 2 | 0.051 | 371 |
| APP | Amyloid Beta Precursor Protein | 0.070 | 366 |
| IRF1 | Interferon Regulatory Factor 1 | 0.036 | 306 |
| STAT3 | Signal Transducer And Activator Of Transcription 3 | 0.049 | 295 |
| TEAD4 | TEA Domain Transcription Factor 4 | 0.042 | 292 |
| RELA | RELA Proto-Oncogene, Nf-Kb Subunit | 0.039 | 278 |
#BC = Betweenness Centrality
$DC = Degree Centrality
Salt Sensitivity Protein Interaction Network (SSPIN)
PIN genes have been grouped into hubs and bottlenecks based on criteria of graph centrality to establish a large network of protein interactions. The cut-off limit for hubs and bottlenecks was defined in the methods section on the basis of Formulas 1 & 2. The degree of the hubs ranged from 86 to 882 nodes which makes an average connectivity of 208 edges per node. We obtained 40 hubs, 502 bottlenecks and 47 SSGs. 15 of 47 SSGs were also found to act as bottleneck in the interactome (Table 2). Hubs, bottlenecks and SSGs were together consisted of 574 genes with 5356 interactions. For the ease of exploration genes in hubs, bottlenecks and SSGs were grouped as HBS. The interaction among these 574 genes in HBS were mapped from PIN to create new network of Salt Sensitivity Protein Interaction Network.
Table 2. The salt sensitivity genes overlapping with bottleneck genes.
| Symbol | Name | BC# | DC$ |
|---|---|---|---|
| ADD1 | Adducin 1 | 0.001 | 17 |
| ADRB2 | Adrenoceptor beta 2 | 0.00305 | 62 |
| AGT | Angiotensinogen | 0.00105 | 16 |
| AGTR1 | Angiotensin II receptor type 1 | 0.00097 | 9 |
| ATP6AP2 | ATPase H+ transporting accessory protein 2 | 0.00102 | 20 |
| CYP11B2 | Cytochrome P450 family 11 subfamily B member 2 | 0.00084 | 4 |
| GNAI2 | G protein subunit alpha i2 | 0.00283 | 31 |
| GNB3 | G protein subunit beta 3 | 0.00041 | 10 |
| MME | Membrane metalloendopeptidase | 0.00047 | 22 |
| NEDD4L | Neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase | 0.00216 | 46 |
| PRCP | Prolylcarboxypeptidase | 0.00052 | 9 |
| PREP | Prolyl endopeptidase | 0.00048 | 11 |
| SCNN1A | Sodium channel epithelial 1 alpha subunit | 0.00043 | 12 |
| SGK1 | Serum/glucocorticoid regulated kinase 1 | 0.00149 | 32 |
| WNK1 | WNK lysine deficient protein kinase 1 | 0.00096 | 33 |
#BC = Betweenness Centrality
$DC = Degree Centrality
Functional enrichment analysis
We used the ToppGene computational annotation system to determine the functional and biological importance of the genes. The genes of HBS have been enriched by 2192 biological processes (BP), 210 molecular functions (MF), 246 cellular components (CC), 642 pathways and 1669 diseases. Analysis of enrichment accounted for about 125 obesity genes. The obesity related genes consisted of 23 SSGs, 16 hubs and 84 bottleneck genes. Approximately 50% of the SSGs were observed to be involved in obesity via functional enrichment analysis (Table 3). These genes were also involved in pathways associated with obesity like regulation of lipolysis in adipocytes, adipogenesis, adipocytokine signaling pathway, renin-angiotensin system, signaling by leptin, toll-like receptor pathway, PI3K-Akt signaling pathway, ras signaling pathway, cytokine signaling in immune system insulin pathway, glucocorticoid receptor regulatory network and NF-kappa B signaling pathway (Table 4). The detailed list of genes involved in these pathways are given in the S3 Table.
Table 3. The genes involved in obesity categorized as hubs, bottlenecks and salt sensitivity genes.
| Category | Genes | P-value |
|---|---|---|
| Salt sensitivity genes | ACE, ACE2, ADD1, ADRB2, AGT, AGTR1, AGTR2, ANPEP, ATP6AP2, CMA1, CYP17A1, GNB3, GRK4, KLK1, LNPEP, MAS1, MME, NEDD4L, PRCP, PRKG1, REN, SGK1, TH | 2.52 x 10−16 |
| Hubs | EGR1, JUND, FOS, APP, STAT3, JUN, STAT1, ATF3, SIRT7, FOXM1, TBL1XR1, BAG3, HSPB1, CEBPD, HNRNPA1, VCAM1 | 2.52 x 10−16 |
| Bottlenecks | CALM1, PCNA, JUNB, CRK, SHC1, GAPDH, WWOX, ITCH, HSPA1A, CRY2, NFKB1, MLH1, PKM, HSPD1, PTPN11, MAP1LC3B, TUFM, APC, SNRNP200, CDK5, CALR, HLA-C, GTF2I, PRKAR1A, BCL2L1, TNF, IGF1R, ZFP36, NR4A1, TANK, SOD2, KRT18, JAK3, SMARCA2, NUP62, PRKCZ, DNMT1, ATG5, DNAJB1, STAT5B, LEPR, VDR, PIK3CA, PPIA, FOXO3, MYD88, CAST, DDAH2, VEGFA, SOCS3, PINK1, COL1A1, THBS1, ACAT2, THRA, SNAP29, VTI1B, PER1, TPI1, RGS2, BMPR1A, NPHP1, FTL, GTF2H1, APOE, CYCS, ABCA1, CSK, TIMM44, GNAQ, C3, POLDIP2, SLU7, ST13, COL4A1, LAMA1, SDC2, IGF1, BGN, CFH, ADM, WASF1, HGF, C1QTNF6 | 2.52 x 10−16 |
Table 4. The enriched pathways that are closely associated with obesity or obesity related diseases.
| Pathway | P-value | Source | Gene’s Count |
|---|---|---|---|
| Adipocytokine signaling pathway | 1.86E-02 | KEGG | 7 |
| Adipogenesis | 5.56E-03 | Wikipathways | 12 |
| Cellular responses to stress | 7.36E-06 | REACTOME | 39 |
| Chemokine signaling pathway | 4.22E-04 | KEGG2 | 18 |
| Cytokine Signaling in Immune system | 9.58E-08 | REACTOME | 62 |
| Glucocorticoid receptor regulatory network | 1.45E-07 | PID | 16 |
| Hemostasis | 7.39E-04 | REACTOME | 43 |
| Insulin signaling pathway | 6.39E-05 | PID1 | 9 |
| Interferon Signaling | 2.18E-06 | REACTOME | 24 |
| MAPK signaling pathway | 6.88E-04 | KEGG | 22 |
| Mineralocorticoid biosynthesis | 4.78E-03 | BIOCYC | 2 |
| NF-KB signaling pathway | 1.12E-03 | BioCarta | 5 |
| NOD-like receptor signaling pathway | 5.32E-04 | KEGG | 17 |
| PI3K-Akt signaling pathway | 8.92E-06 | KEGG | 32 |
| Ras signaling pathway | 3.61E-04 | KEGG | 21 |
| Regulation of lipolysis in adipocytes | 1.16E-03 | KEGG | 8 |
| Renin secretion | 2.61E-04 | KEGG | 10 |
| Renin-angiotensin system | 3.25E-30 | KEGG | 22 |
| Signaling by Leptin | 8.10E-03 | REACTOME | 19 |
| Signaling by Rho GTPases | 2.63E-03 | REACTOME | 30 |
| Sodium/Calcium exchangers | 7.92E-03 | Reactome | 3 |
| Sphingolipid signaling pathway | 2.94E-03 | KEGG | 12 |
| TNF signaling pathway | 1.46E-06 | KEGG | 17 |
| Toll-like receptor pathway | 2.23E-03 | BioCarta | 6 |
| Type I diabetes mellitus | 2.61E-02 | KEGG | 5 |
| Type II diabetes mellitus | 8.93E-03 | KEGG | 6 |
1PID = pathway interaction database
2KEGG = Kyoto Encyclopedia of Genes and Genomes
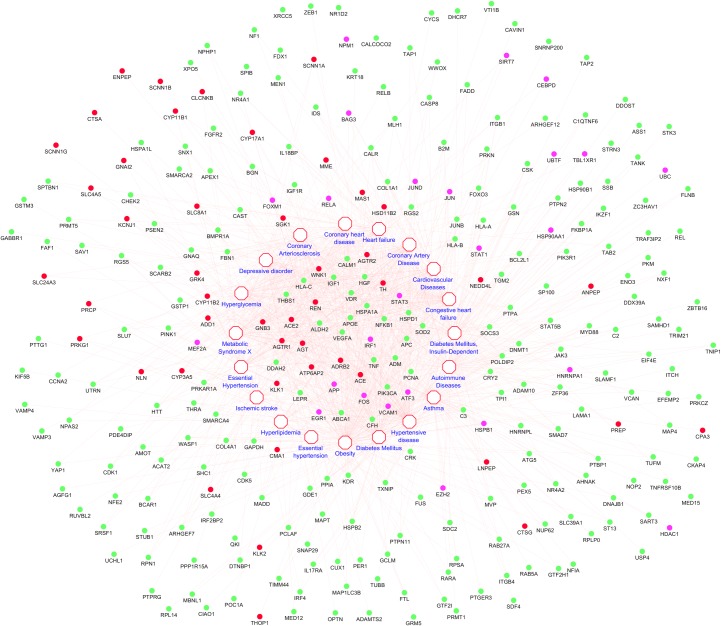
The enriched genes were also involved in obesity related diseases like Diabetes Mellitus, Hypertensive disease, Asthma, Autoimmune Diseases, Diabetes Mellitus (Insulin-Dependent), Congestive heart failure, Cardiovascular Diseases, Coronary Artery Disease, Heart failure, Coronary heart disease, Coronary Arteriosclerosis, Depressive disorder, Hyperglycemia, Metabolic Syndrome X, Essential Hypertension, Ischemic stroke, Hyperlipidemia. Obesity is one of leading cause of aforesaid diseases. The interaction map of genes to the diseases is depicted in the Fig 2.
Fig 2. The interaction map of disease to genes.
The red nodes represents salt sensitivity genes, pink and green nodes represents hubs and bottlenecks respectively.
Co-expression analysis
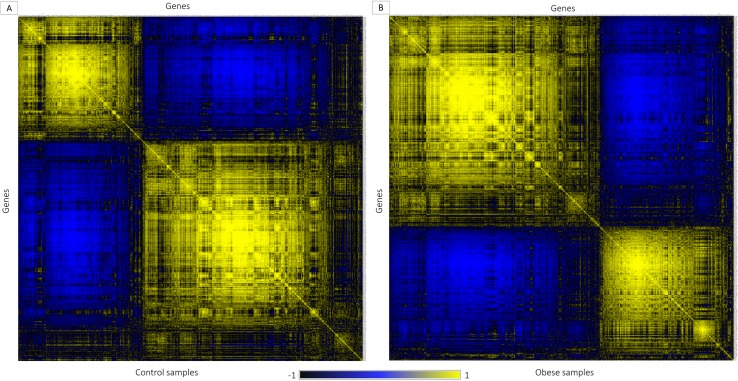
The expression pattern similarity between 574 HBS genes was established and ranked based on Pearson's correlation algorithm (Fig 3) for array of control and disease samples. For control and disease samples (Formula 3), the algorithm created PCC for 328329 pair of genes from 574 genes. Gene pairs were screened in this approach based on established concepts such as i) gene expression level with high positive correlation. ii) Genes with similar patterns of speech are more likely to interact. In obesity studies, gene pairs with value r = 0.8 are chosen from the correlation map as higher r score indicates a greater relationship. Corresponding gene pairs were extracted from normal correlation map to identify the variation in the co-expression from obesity to normal sample. Totally, 226 genes are observed to co-express with obesity related genes with 1126 interactions in obesity condition (Fig 4). There were 88 obesity related genes and 23 SSGs in the set which were co-expressed in samples of obese adipose tissue. We focused on the 23 SSGs that are found to have co-expressed with obesity related genes.
Fig 3. Representation of gene-gene correlation plot.
The correlation plots illustrate substantial variations in gene expression among the gene pairs in the control (lean) and obese samples. A). Gene-gene correlation of lean samples (control), B). Gene-gene correlation of obese samples (disease)
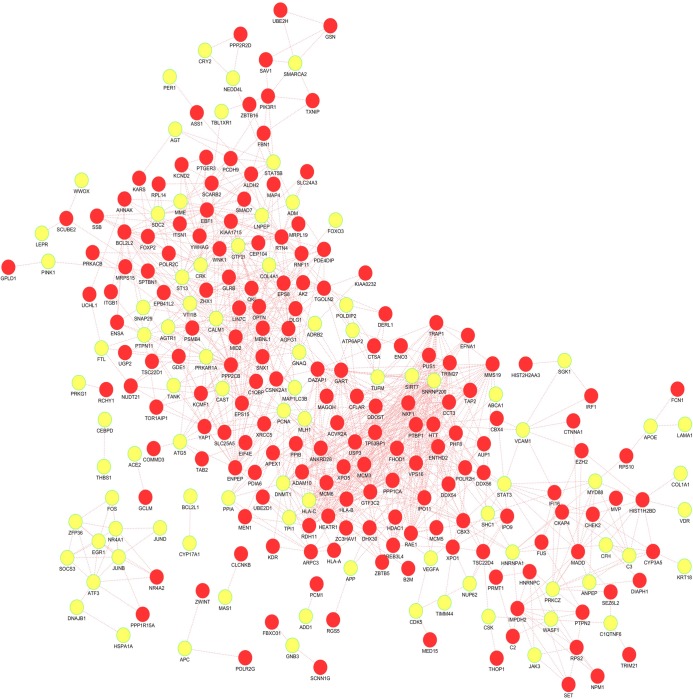
Fig 4. The plot of genes co-expressed with obesity related genes.
The obese condition where yellow nodes represents obesity related genes.
By performing co-expression analysis, we obtained 23 co-expressed SSGs with obesity related genes. Eight among the 23 co-expressed genes were not previously reported for the disease obesity via functional enrichment analysis. The list of co-expressed SSGs are depicted in the Table 5. We developed an interaction map of unreported SSGs with obesity related genes (Fig 5) by taking their co-relation score as weight (Table 6). We extracted the edge weight of gene pairs in both obese and normal sample to identify the distinct variations across set of two conditions. This attempt was performed because of the fact that differentially co-expressed genes participate in numerous biological processes resulting in adverse or complementary effects.
Table 5. List of co-expressed salt sensitive genes with their identity in obesity.
| Gene | Name | Role in obesity |
|---|---|---|
| ACE2 | Angiotensin I Converting Enzyme 2 | Reported |
| ADD1 | Adducin 1 | Reported |
| ADRB2 | Adrenoceptor Beta 2 | Reported |
| AGT | Angiotensinogen | Reported |
| AGTR1 | Angiotensin Ii Receptor Type 1 | Reported |
| ANPEP | Alanyl Aminopeptidase, Membrane | Reported |
| ATP6AP2 | Atpase H+ Transporting Accessory Protein 2 | Reported |
| CYP17A1 | Cytochrome P450 Family 17 Subfamily A Member 1 | Reported |
| GNB3 | G Protein Subunit Beta 3 | Reported |
| LNPEP | Leucyl And Cystinyl Aminopeptidase | Reported |
| MAS1 | Mas1 Proto-Oncogene, G Protein-Coupled Receptor | Reported |
| MME | Membrane Metalloendopeptidase | Reported |
| NEDD4L | Neural Precursor Cell Expressed, Developmentally Down-Regulated 4-Like, E3 Ubiquitin Protein Ligase | Reported |
| PRKG1 | Protein Kinase Cgmp-Dependent 1 | Reported |
| SGK1 | Serum/Glucocorticoid Regulated Kinase 1 | Reported |
| CLCNKB | Chloride Voltage-Gated Channel Kb | Unreported |
| CTSA | Cathepsin A | Unreported |
| CYP3A5 | Cytochrome P450 Family 3 Subfamily A Member 5 | Unreported |
| ENPEP | Glutamyl Aminopeptidase | Unreported |
| SCNN1G | Sodium Channel Epithelial 1 Gamma Subunit | Unreported |
| SLC24A3 | Solute Carrier Family 24 Member 3 | Unreported |
| THOP1 | Thimet Oligopeptidase 1 | Unreported |
| WNK1 | Wnk Lysine Deficient Protein Kinase 1 | Unreported |
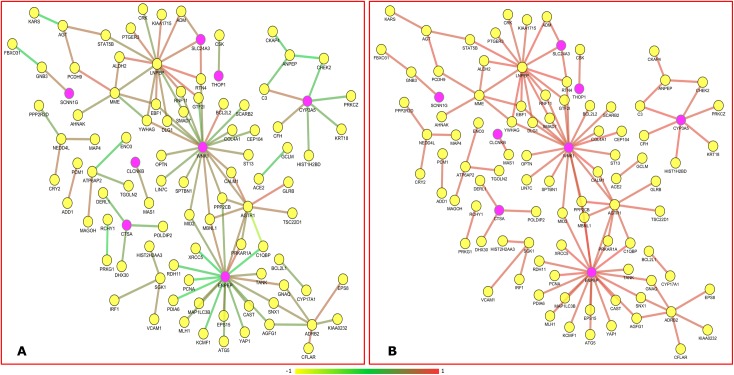
Fig 5. The plot depicts the correlation score of gene pairs in obese and control conditions.
The color scale (-1 to +1) represents the correlation value. Higher the value higher is the correlation. (A) Represents gene-gene correlation in normal samples and (B) represents their corresponding correlation in obese condition. Pink nodes depict novel genes that are co-expressed with obesity related genes in obese condition and Yellow nodes represent obesity related genes.
Table 6. Interactions of unreported salt sensitive genes in obese and normal condition with their corresponding co-relation score as weights.
| Gene-1 | Gene-2 | Obese1 | Normal2 |
|---|---|---|---|
| WNK1 | CALM1 | 0.9439 | 0.5472 |
| ENPEP | C1QBP | 0.9417 | -0.2225 |
| ENPEP | PCNA | 0.9240 | -0.2255 |
| ENPEP | MBNL1 | 0.9156 | 0.5669 |
| WNK1 | CEP104 | 0.9145 | 0.2931 |
| ENPEP | XRCC5 | 0.9110 | -0.1939 |
| CYP3A5 | HIST1H2BD | 0.9040 | 0.2237 |
| CYP3A5 | C3 | 0.9015 | 0.6306 |
| CLCNKB | MAS1 | 0.9001 | 0.4543 |
| ENPEP | CAST | 0.8897 | 0.3039 |
| CYP3A5 | CFH | 0.8787 | 0.5056 |
| CYP3A5 | KRT18 | 0.8696 | 0.1807 |
| ENPEP | SNX1 | 0.8659 | 0.3880 |
| SLC24A3 | RTN4 | 0.8625 | 0.7491 |
| ENPEP | MID2 | 0.8569 | 0.3283 |
| ENPEP | GNAQ | 0.8480 | 0.5793 |
| WNK1 | SPTBN1 | 0.8469 | 0.3412 |
| SLC24A3 | LNPEP | 0.8434 | 0.6549 |
| WNK1 | OPTN | 0.8420 | 0.3940 |
| ENPEP | MAP1LC3B | 0.8410 | 0.4832 |
| WNK1 | ST13 | 0.8377 | 0.4385 |
| WNK1 | LIN7C | 0.8362 | 0.2726 |
| WNK1 | DLG1 | 0.8352 | 0.3487 |
| ENPEP | PPP2CB | 0.8341 | 0.2886 |
| WNK1 | BCL2L2 | 0.8338 | 0.7179 |
| ENPEP | AGFG1 | 0.8333 | 0.2477 |
| SLC24A3 | ADM | 0.8324 | 0.5586 |
| ENPEP | MLH1 | 0.8309 | 0.2835 |
| WNK1 | RNF11 | 0.8297 | 0.3692 |
| CTSA | POLDIP2 | 0.8285 | 0.1705 |
| CTSA | DERL1 | 0.8279 | 0.1225 |
| ENPEP | YAP1 | 0.8258 | 0.1928 |
| WNK1 | SMAD7 | 0.8237 | 0.2519 |
| ENPEP | KCMF1 | 0.8225 | -0.0395 |
| WNK1 | YWHAG | 0.8217 | 0.5680 |
| SCNN1G | GNB3 | 0.8216 | 0.5586 |
| ENPEP | PRKAR1A | 0.8215 | 0.7386 |
| CYP3A5 | PRKCZ | 0.8213 | 0.1144 |
| ENPEP | RDH11 | 0.8198 | -0.0770 |
| WNK1 | COL4A1 | 0.8191 | 0.3485 |
| CTSA | DHX30 | 0.8171 | 0.2648 |
| WNK1 | EBF1 | 0.8168 | 0.3779 |
| ENPEP | ATG5 | 0.8151 | 0.2386 |
| ENPEP | AGTR1 | 0.8149 | 0.4997 |
| ENPEP | EPS15 | 0.8134 | 0.5767 |
| WNK1 | AGTR1 | 0.8124 | 0.4890 |
| WNK1 | MBNL1 | 0.8123 | 0.5277 |
| WNK1 | RTN4 | 0.8121 | 0.4261 |
| ENPEP | PDIA6 | 0.8111 | -0.2465 |
| CYP3A5 | CHEK2 | 0.8098 | 0.0108 |
| WNK1 | PPP2CB | 0.8077 | 0.4375 |
| WNK1 | SCARB2 | 0.8059 | 0.1818 |
| THOP1 | CSK | 0.8058 | 0.3290 |
| WNK1 | MID2 | 0.8047 | 0.3940 |
| ENPEP | TANK | 0.8027 | 0.3945 |
| WNK1 | GTF2I | 0.8001 | 0.1125 |
1obese = correlation score in obese sample
2normal = correlation score in normal sample
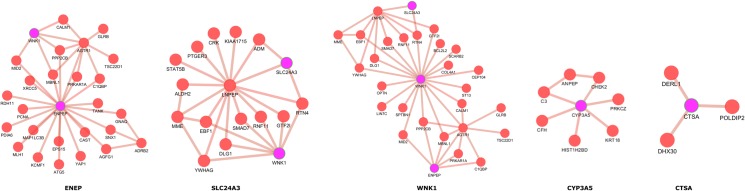
It is very clear from the plot that majority of the co-expressed genes in the obese conditions are not co-expressed in normal conditions. Considering them as a disease subnetwork, we calculated the local topological parameters based on graph theory. Among 8 unreported SSGs, the highly connected genes with obesity related genes is ENPEP followed by WNK1. These two genes were having 21 and 20 direct connectivity to the obesity related gene in the co-expressed state. The SSGs, THOP1, CLCNKB, SCNN1G and THOP1 were having poor connectivity in the disease subnetwork. Notably, CYP3A5 and CTSA formed two separate networks with connectivity 6 and 3 respectively to obesity related gene. The interactions of the unreported SSGs with obesity related genes was separated and depicted in the Fig 6. We have narrowed down unreported SSGs to 5 prioritized genes (ENPEP, WNK1, CYP3A5, SLC24A3 and CTSA) based on their co-expression and topological parameters.
Fig 6. The partners of prioritized unreported salt sensitive genes (ENPEP, WNK1, CYP3A5, SLC24A3 and CTSA) which are co-expressed with obesity related genes in obese condition.
An attempt was made to associate novel genes found in this study to the genome wide association studies on many disease traits from around the world in the GWAS catalog (MacArthur et al., 2016). We extracted the reported traits of these co-expressed genes from GWAS catalog to identify their association to obesity (Table 7). Many of these traits were related to obesity or its associated traits in cardiovascular or metabolic diseases.
Table 7. The traits extracted from GWAS catalogue for the unreported genes co-expressed with obesity related genes.
| Gene | Trait | PubMed |
|---|---|---|
| ENPEP | atrial fibrillation | 17603472 |
| ENPEP | systolic blood pressure | 21572416 |
| ENPEP | diastolic blood pressure | 21572416 |
| ENPEP | cognitive impairment, cognitive decline measurement | 26252872 |
| ENPEP | eye color | 29109912 |
| ENPEP | lung carcinoma | 28604730 |
| ENPEP | metabolite measurement | 21886157 |
| ENPEP | pursuit maintenance gain measurement | 29064472 |
| SLC24A3 | chronic obstructive pulmonary disease, smoking initiation | 21685187 |
| SLC24A3 | fat body mass | 28224759 |
| SLC24A3 | matrix metalloproteinase measurement | 20031604 |
| SLC24A3 | mean platelet volume | 27863252 |
| SLC24A3 | migraine disorder | 27182965 |
| SLC24A3 | Psychosis | 24132900 |
| SLC24A3 | FEV/FEC ratio | 22424883 |
| SLC24A3 | pulse pressure measurement | 28135244 |
| SLC24A3 | unipolar depression, response to escitalopram, response to citalopram, mood disorder | 27622933 |
| SLC24A3 | Age at smoking initiation in chronic obstructive pulmonary disease | 21685187 |
| SLC24A3 | Daytime sleepiness | 28604731 |
| SLC24A3 | Matrix metalloproteinase levels | 20031604 |
| SLC24A3 | Migraine | 27182965 |
| SLC24A3 | Pulmonary function decline | 22424883 |
| SLC24A3 | Pulse pressure | 28135244 |
| SLC24A3 | QT interval | 27958378 |
| WNK1 | body mass index | 25673413 |
| WNK1 | colorectal cancer | 24836286 |
| WNK1 | eosinophil count | 27863252 |
| WNK1 | eosinophil percentage of leukocytes | 27863252 |
| WNK1 | lung carcinoma | 28604730 |
| WNK1 | smoking status measurement, lung carcinoma | 28604730 |
| WNK1 | squamous cell lung carcinoma | 28604730 |
| WNK1 | blood manganese measurement | 26025379 |
| WNK1 | Malignant epithelial tumor of ovary, response to paclitaxel | 29367611 |
| WNK1 | Stroke | 19369658 |
| CYP3A5 | Blood metabolite levels | 25898920 |
| CYP3A5 | Borderline personality disorder | 28632202 |
| CYP3A5 | Cognitive decline rate in late mild cognitive impairment | 26252872 |
| CYP3A5 | Disease progression in age-related macular degeneration | 29346644 |
| CYP3A5 | Early childhood aggressive behavior | 26087016 |
| CYP3A5 | Factor VII | 17903294 |
| CYP3A5 | Obesity-related traits | 23251661 |
| CYP3A5 | Ticagrelor levels in individuals with acute coronary syndromes treated with ticagrelor | 25935875 |
| CYP3A5 | Blood metabolite ratios | 24816252 |
Discussion
Traditional gene profiling approaches are based on detecting individual targeted genes showing variations in the experimental group versus the control one. However, mere identification of differentially expressed genes cannot always help in understanding biological pathways (metabolism, transcription, and gene interactions, among others) regulations involved in the disease pathogenesis [35]. This is especially true in case of multifaceted or complex disorders like obesity, which do not progress because of instabilities in a single gene, but due to the changes in several pathways comprising of various biological networks [14]. In the current study, we investigated the concepts of gene regulatory networks in order to profile the significant variations of salt-sensitive genes involved in obesity.
Local parameter DC and global parameter BC were used to dissect the complex interactome. DC of a gene is the number of partners that are connected to that specific gene. Protein Interaction Network (PIN) are mathematical representations of physical and/or functional interaction between nodes, where nodes are the genes and the edges represent the connection between them, which may be binding possibility, metabolic interaction or regulatory crosstalks [36]. In our built PIN, significant alterations were observed in the expression level of h selected genes in our experimental settings. Initially, a complex network of significant genes from adipose tissue was constructed which was further decomposed to a Salt Sensitivity Protein Interaction Network based on hubs and bottlenecks. Hubs are considered as key features in networks, because they project critical intersections, which gets disturbs the networks whenever they are removed [37]. In the constructed interactome PIN, highly essential genes show high degree of connectivity. Several publications strongly suggested that diseased genes have higher connectivity and cross-talks when compared to non-diseased ones which are supporting hubs impact in the network [38]. We obtained 40 hubs with an average connectivity of 208 edges. The enrichment analysis revealed that 16 hub genes were involved in obesity and 13 hubs were involved in Type 2 Diabetes, closely related to obesity. Thus, the identification of hub molecules in the PIN is of substantial interest to get better insights of the disease pathogenesis. On other hand, functionally relevant vertices (nodes) in the network were detected using betweenness centrality (BC). In fact, This approach helped to sort-out vertices linking dense networks, rather than nodes located inside the dense cluster[28]. Functional enrichment analysis represented 84 bottleneck genes in obesity.
The unintended interactions of the genes may lead to deregulated functions. Hence, to better understand the gene function in cellular context, we need to understand how genes are interconnected together within several biological processes and molecular signaling pathways. In fact this type of structural and functional bio interactome can be created by evaluating the functional features of the genes. Therefore, carrying out gene enrichment analysis is a vital part in exploring the high-throughput data extracted from different biological observations and experiments. This methodology helps to discover the non-predefined interaction between functional genes that significantly regulate different biological. Gene ontology analysis depicted the involvement of 125 genes in obesity and 24 genes among them were SSGs contributing to 50 percentage of total SSGs. These findings signifies the critical role of SSGs in the role of obesity. To explore more on salt related genes co-expression analysis of obesity related genes in adipose tissue was carried out. By performing co-expression analysis, we obtained 23 co-expressed SSGs with obesity related genes. Eight among the 23 co-expressed genes were not previously reported for the disease obesity via gene ontology analysis. Gene co-correlation can be explained by the fact that genes showing similar regulation/ expression patterns are frequently interconnected together than with arbitrary genes [9]. Interaction map of the unreported SSGs with obesity related genes showed stronger interactions in disease state. It is very clear from the plot (Fig 5) that majority of the co-expressed genes in the obese conditions are not co-expressed in normal conditions. The novel obesity associated SSG and their interactions supports the view that the differentially co-expressed genes are likely to get involved in numerous molecular processes resulting in adverse or balancing effects [39].
The established theory in network biology is that disease related genes existing in close physical proximity are most likely to cause diseases with similar molecular basis. In addition, in a network of disease genes, the non-disease genes are identified to have a higher tendency to interact with other disease genes [40]. Considering the theory, we looked into disease and pathway related to the prioritized gene from unreported SSGs. WNK1 and ENPEP act as central hub in the network with high number co-expressed partners. In the functional enrichment data, the gene WNK1 is reported in diseases like Diabetes Mellitus, Cardiovascular Diseases, Metabolic Syndrome X, Hyperglycemia and heart failure. These enriched diseases also show close relationship with obesity. Recent report by Ding et al., [41] in mouse model suggests WNK1 as a novel signaling molecule involved in development of obesity. It suggests lack of Akt3 in adipocytes rises the WNK1 protein level which in turn activates SGK1 and stimulates adipogenesis through phosphorylation and inhibition of FOXO1 transcription factor, subsequently, activating the transcription of PPARg in adipocytes. Increased adipocyte results in high fat accumulation and ultimately to obesity. Thus, WNK1, can act as one of the potential biomarker or targets for controlling obesity. Additionally, at pathway level, WNK1 is known to be a potent regulator of Na+ and Cl- ions transport, and consequently the blood pressure. Ewout et al, (2011) describes about the role of WNKs in salt metabolism via regulating sodium, chlorine, potassium and blood pressure [42]. WNKs are involved in crucial molecular pathways via connecting hormones such as angiotensin II and aldosterone to sodium and potassium transport. WNK1 is significantly involved in homeostasis and several biological processes regulations including and not limited to cell survival, proliferation and signaling fates. WNK1 activates sodium channel epithelial (ENaC) gene subunits SCNN1A, SCNN1B, and SCNN1D. It is also known as an activator of SGK1. In fact, by inhibiting WNK4 activity through kinase phosphorylation, WNK1 controls Na+ and Cl- ions transport. Moreover, WNK1 plays a switch role-like (activation/inhibition) of the Na-K-Cl cotransporters (NKCC) respectively [43].
ENPEP is a member of the M1 family of endopeptidases. It is plays a role in the catabolic pathway of the renin angiotensin system which in turn is involved in regulation of blood pressure [44]. The gene is observed in Hypertensive disease which are closely associated with obesity. Currently, inhibition of ENPEP activity is one of procedure used to treat hypertension condition. Hypertension is a growing problem affecting 40% percent of adults due to the growing prevalence of obesity and diabetes in many parts of the world [45]. In addition, DNA methylation study in human adipose tissue reveals ENPEP as one of the differentially methylated genes associated with obesity and related traits [46]. ENPEP is found to be a candidate gene associated with obesity and hypertension traits in GWAS (Genome Wide Association study) studies. ENPEP is highly correlated with obesity related genes and also correlated with the diseases that may be comorbidity conditions of obesity. Therefore, our work provides strong evidence for ENPEP to be a novel gene that contributing to obesity.
CYP3A5 plays a role in the metabolism of many drugs and other metbolites, such as steroids. CYP3A5 is also involved in the oxidative metabolism of xenobiotics, as well as calcium channel blocking drugs and immunosuppressive drugs. CYP3A5 is a member of the cytochrome P450 superfamily of enzymes. These proteins are monooxygenases catalyzing reactions in metabolism of drugs, cholesterol, steroids and other lipids. The main functions associated with CYP3A5 are monooxygenase activity, iron ion binding, lipid metabolism and oxidoreductase activity [47].
Potassium-dependent sodium/calcium exchanger (SLC24A3) plays an important role in intracellular calcium homeostasis. It facilitates exchange of intracellular Ca++ and K+ ions for extracellular sodium ions [48]. CTSA is a member of cathepsins family which are a group of lysosomal proteases that have a key role in cellular protein turnover. CTSA is not directly reported in obesity, but an analysis performed by Nadia et al., (2010) implicates cysteine proteases cathepsins S, L, and K in complications of obesity [49]. Similarly, a study conducted by Araujo et al., (2018) reports CTSB, a member in Cathepsin family, controls autophagy in adipocytes. In obese individuals, the expression of this gene increases which in turn regulates inflammatory markers [50]. In our analysis CTSA is co-expressed with obesity related genes suggesting a critical role in the pathway of obesity since the members of Cathespin family plays import role in obesity. The major functions associated with CTSA are glycosphingolipid metabolism, protein transport and enzyme activating activity.
In GWAS analysis, the genes CYP3A5, SLC24A3 and CTSA are observed in obesity related diseases like Hypertensive disease, Asthma, Coronary Artery Disease, Essential Hypertension, Hypertensive disease and Heart failure. We found the gene CYP3A5 is reported as one of loci associated with obesity related traits in GWAS studies [51]. It is also associated with Factor VII and blood metabolite levels. Recent study by Takahashi et al., [52] reports the relationship of factor VII and obesity. The results propose Factor VII is an adipokine, enhanced by TNF-α or isoproterenol, which plays crucial role in the pathogenesis of obesity. SLC24A3 and WNK1 are mapped to traits like fat body mass and body mass index which are closely associated with obesity. This analysis of integrating GWAS studies also substantiates the possible association of novel genes identified through this study to obesity related traits and comorbidity symptoms and diseases.
We acknowledge that our strategy has some technical constraints. First, since experimentally derived protein interactions were retrieved using Bisogenet plugin. This plugin employs multiple databases of protein-protein interactions hence any interaction which has not been updated in those databases may not have been included in our study. In addition to this the insufficiency of data pertaining to certain genes in the Gene Ontology (GO) should also be considered. In order to overcome these limitations, we tried to include protein interaction based on co-expression. Overall, our research analysis has presented the effectiveness of linking genetic expression with their functional relationship in identification of obesity candidate genes. In order to demonstrate the involvement of the novel candidate genes mentioned in this study further experimental validation is required.
Conclusions
This work systematically outlines an integrated bioinformatics pipeline for figuring out the most indispensable key signatures from the interactome Salt Sensitivity Protein Interaction Network (SSPIN). The findings with biological relevance depict 50% of the SSGs have experimental evidences for their role in the pathogenesis of obesity. A detailed parametric downstream analysis based on biological insights, illustrated 5 candidate genes that can act as potential biomarker or target for obesity. To authenticate our results, we illustrate the possible role of ENPEP and WNK1 which appeared in the top prioritized list. Overall, our research analysis has presented the effectiveness of linking genetic expression with their functional relationship in identification of obesity candidate genes.
Supporting information
(PDF)
(PDF)
(XLSX)
Acknowledgments
This project was funded by the Deanship of Scientific Research (DSR) at King Abdulaziz University, under grant no. HiCi-63-130-35. The authors are thankful to DSR for the technical and the financial support.
Abbreviations
- BC
Betweenness Centrality
- BIND
Biomolecular Interaction Network Database
- BioGRID
Biological General Repository for Interaction Datasets
- DC
Degree Centrality
- DEGs
differentially expressed genes
- DIP
Database of Interacting Proteins
- FDR
False Discovery Rate
- GWAS
Genome-wide association studies
- HPRD
Human Protein Reference Database
- MINT
The Molecular Interaction database
- PCC
Pearson’s correlation algorithm
- PPIM
Protein Interaction Map
- RMA
Robust Multiarray Average
- SSG
Salt sensitivity genes
- SSPIN
Salt Sensitivity Protein Interaction Network
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This project was funded by the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, KSA, under grant no. HiCi-63-130-35. The authors are thankful to DSR for the technical and the financial support.
References
- 1.Jousheghany F, Phelps J, Crook T, Hakkak R. Relationship between level of HbA1C and breast cancer. BBA Clin. 2016;6:45–8. Epub 2016/12/14. 10.1016/j.bbacli.2016.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanti JF, Ceppo F, Jager J, Berthou F. Implication of inflammatory signaling pathways in obesity-induced insulin resistance. Front Endocrinol (Lausanne). 2012;3:181 Epub 2013/01/15. 10.3389/fendo.2012.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gabrielli AP, Manzardo AM, Butler MG. Exploring genetic susceptibility to obesity through genome functional pathway analysis. Obesity (Silver Spring). 2017;25(6):1136–43. Epub 2017/05/06. 10.1002/oby.21847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iyer A, Fairlie DP, Prins JB, Hammock BD, Brown L. Inflammatory lipid mediators in adipocyte function and obesity. Nat Rev Endocrinol. 2010;6(2):71–82. Epub 2010/01/26. 10.1038/nrendo.2009.264 . [DOI] [PubMed] [Google Scholar]
- 5.Mishra S, Ingole S, Jain R. Salt sensitivity and its implication in clinical practice. Indian Heart J. 2018;70(4):556–64. Epub 2018/09/02. 10.1016/j.ihj.2017.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rassler B. The Renin-Angiotensin System in the Development of Salt-Sensitive Hypertension in Animal Models and Humans. Pharmaceuticals (Basel). 2010;3(4):940–60. Epub 2010/03/29. 10.3390/ph3040940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunner N, Quezada C, Berndt FA, Canovas J, Rojas CV. Angiotensin II signaling in human preadipose cells: participation of ERK1,2-dependent modulation of Akt. PLoS One. 2013;8(10):e75440 Epub 2013/10/08. 10.1371/journal.pone.0075440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bader S, Kuhner S, Gavin AC. Interaction networks for systems biology. FEBS Lett. 2008;582(8):1220–4. Epub 2008/02/20. 10.1016/j.febslet.2008.02.015 . [DOI] [PubMed] [Google Scholar]
- 9.Grigoriev A. A relationship between gene expression and protein interactions on the proteome scale: analysis of the bacteriophage T7 and the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 2001;29(17):3513–9. Epub 2001/08/28. 10.1093/nar/29.17.3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clough E, Barrett T. The Gene Expression Omnibus Database. Methods Mol Biol. 2016;1418:93–110. Epub 2016/03/24. 10.1007/978-1-4939-3578-9_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26(19):2363–7. Epub 2010/08/07. 10.1093/bioinformatics/btq431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47 Epub 2015/01/22. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown L. The Conditional Level of Student's $t$ Test. Ann Math Statist. 1967;38(4):1068–71. 10.1214/aoms/1177698776 [DOI] [Google Scholar]
- 14.Sabir JSM, El Omri A, Shaik NA, Banaganapalli B, Al-Shaeri MA, Alkenani NA, et al. Identification of key regulatory genes connected to NF-kappaB family of proteins in visceral adipose tissues using gene expression and weighted protein interaction network. PLoS One. 2019;14(4):e0214337 Epub 2019/04/24. 10.1371/journal.pone.0214337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oughtred R, Stark C, Breitkreutz BJ, Rust J, Boucher L, Chang C, et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47(D1):D529–D41. Epub 2018/11/27. 10.1093/nar/gky1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32(Database issue):D449–51. Epub 2003/12/19. 10.1093/nar/gkh086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13(10):2363–71. Epub 2003/10/04. 10.1101/gr.1680803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohart F, Eslami A, Matigian N, Bougeard S, Le Cao KA. MINT: a multivariate integrative method to identify reproducible molecular signatures across independent experiments and platforms. BMC Bioinformatics. 2017;18(1):128 Epub 2017/03/01. 10.1186/s12859-017-1553-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, et al. IntAct—open source resource for molecular interaction data. Nucleic Acids Res. 2007;35(Database issue):D561–5. Epub 2006/12/06. 10.1093/nar/gkl958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. Epub 2003/11/05. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Assenov Y, Ramirez F, Schelhorn SE, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24(2):282–4. Epub 2007/11/17. 10.1093/bioinformatics/btm554 . [DOI] [PubMed] [Google Scholar]
- 22.Shaik NA, Banaganapalli B. Computational Molecular Phenotypic Analysis of PTPN22 (W620R), IL6R (D358A), and TYK2 (P1104A) Gene Mutations of Rheumatoid Arthritis. Front Genet. 2019;10:168 Epub 2019/03/23. 10.3389/fgene.2019.00168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaik NA, Al-Kreathy HM, Ajabnoor GM, Verma PK, Banaganapalli B. Molecular designing, virtual screening and docking study of novel curcumin analogue as mutation (S769L and K846R) selective inhibitor for EGFR. Saudi J Biol Sci. 2019;26(3):439–48. Epub 2019/03/23. 10.1016/j.sjbs.2018.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaik NA, Awan ZA, Verma PK, Elango R, Banaganapalli B. Protein phenotype diagnosis of autosomal dominant calmodulin mutations causing irregular heart rhythms. J Cell Biochem. 2018;119(10):8233–48. Epub 2018/06/23. 10.1002/jcb.26834 . [DOI] [PubMed] [Google Scholar]
- 25.George G, Valiya Parambath S, Lokappa SB, Varkey J. Construction of Parkinson's disease marker-based weighted protein-protein interaction network for prioritization of co-expressed genes. Gene. 2019;697:67–77. Epub 2019/02/19. 10.1016/j.gene.2019.02.026 . [DOI] [PubMed] [Google Scholar]
- 26.Rakshit H, Rathi N, Roy D. Construction and analysis of the protein-protein interaction networks based on gene expression profiles of Parkinson's disease. PLoS One. 2014;9(8):e103047 Epub 2014/08/30. 10.1371/journal.pone.0103047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lahiri C, Pawar S, Sabarinathan R, Ashraf MI, Chand Y, Chakravortty D. Interactome analyses of Salmonella pathogenicity islands reveal SicA indispensable for virulence. J Theor Biol. 2014;363:188–97. Epub 2014/08/17. 10.1016/j.jtbi.2014.08.013 . [DOI] [PubMed] [Google Scholar]
- 28.Yoon J, Blumer A, Lee K. An algorithm for modularity analysis of directed and weighted biological networks based on edge-betweenness centrality. Bioinformatics. 2006;22(24):3106–8. Epub 2006/10/25. 10.1093/bioinformatics/btl533 . [DOI] [PubMed] [Google Scholar]
- 29.Freeman LC. A Set of Measures of Centrality Based on Betweenness. Sociometry. 1977;40(1):35–41. 10.2307/3033543 [DOI] [Google Scholar]
- 30.Schweda F. Salt feedback on the renin-angiotensin-aldosterone system. Pflugers Arch. 2015;467(3):565–76. Epub 2014/12/17. 10.1007/s00424-014-1668-y . [DOI] [PubMed] [Google Scholar]
- 31.Sanada H, Jones JE, Jose PA. Genetics of salt-sensitive hypertension. Curr Hypertens Rep. 2011;13(1):55–66. Epub 2010/11/09. 10.1007/s11906-010-0167-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freitas SRS. Molecular Genetics of Salt-Sensitivity and Hypertension: Role of Renal Epithelial Sodium Channel Genes. Am J Hypertens. 2018;31(2):172–4. Epub 2017/10/19. 10.1093/ajh/hpx184 . [DOI] [PubMed] [Google Scholar]
- 33.Citterio L, Simonini M, Zagato L, Salvi E, Delli Carpini S, Lanzani C, et al. Genes involved in vasoconstriction and vasodilation system affect salt-sensitive hypertension. PLoS One. 2011;6(5):e19620 Epub 2011/05/17. 10.1371/journal.pone.0019620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server issue):W305–11. Epub 2009/05/26. 10.1093/nar/gkp427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghazalpour A, Doss S, Sheth SS, Ingram-Drake LA, Schadt EE, Lusis AJ, et al. Genomic analysis of metabolic pathway gene expression in mice. Genome Biol. 2005;6(7):R59 Epub 2005/07/07. 10.1186/gb-2005-6-7-r59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albert R. Scale-free networks in cell biology. J Cell Sci. 2005;118(Pt 21):4947–57. Epub 2005/10/29. 10.1242/jcs.02714 . [DOI] [PubMed] [Google Scholar]
- 37.Khosravi P, Gazestani VH, Asgari Y, Law B, Sadeghi M, Goliaei B. Network-based approach reveals Y chromosome influences prostate cancer susceptibility. Comput Biol Med. 2014;54:24–31. Epub 2014/09/10. 10.1016/j.compbiomed.2014.08.020 . [DOI] [PubMed] [Google Scholar]
- 38.Csermely P, Korcsmaros T, Kiss HJ, London G, Nussinov R. Structure and dynamics of molecular networks: a novel paradigm of drug discovery: a comprehensive review. Pharmacol Ther. 2013;138(3):333–408. Epub 2013/02/07. 10.1016/j.pharmthera.2013.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu CL, Juan HF, Huang HC. Functional Analysis and Characterization of Differential Coexpression Networks. Sci Rep. 2015;5:13295 Epub 2015/08/19. 10.1038/srep13295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chavali S, Barrenas F, Kanduri K, Benson M. Network properties of human disease genes with pleiotropic effects. BMC Syst Biol. 2010;4:78 Epub 2010/06/08. 10.1186/1752-0509-4-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding L, Zhang L, Biswas S, Schugar RC, Brown JM, Byzova T, et al. Akt3 inhibits adipogenesis and protects from diet-induced obesity via WNK1/SGK1 signaling. JCI Insight. 2017;2(22). Epub 2017/12/05. 10.1172/jci.insight.95687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoorn EJ, Nelson JH, McCormick JA, Ellison DH. The WNK kinase network regulating sodium, potassium, and blood pressure. J Am Soc Nephrol. 2011;22(4):605–14. Epub 2011/03/26. 10.1681/ASN.2010080827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hadchouel J, Delaloy C, Jeunemaitre X. [WNK1 and WNK4, new players in salt and water homeostasis]. Med Sci (Paris). 2005;21(1):55–60. Epub 2005/01/11. 10.1051/medsci/200521155 . [DOI] [PubMed] [Google Scholar]
- 44.Holmes RS, Spradling-Reeves KD, Cox LA. Mammalian Glutamyl Aminopeptidase Genes (ENPEP) and Proteins: Comparative Studies of a Major Contributor to Arterial Hypertension. J Data Mining Genomics Proteomics. 2017;8(2). Epub 2017/01/01. 10.4172/2153-0602.1000211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao J, Marc Y, Iturrioz X, Leroux V, Balavoine F, Llorens-Cortes C. A new strategy for treating hypertension by blocking the activity of the brain renin-angiotensin system with aminopeptidase A inhibitors. Clin Sci (Lond). 2014;127(3):135–48. Epub 2014/04/05. 10.1042/CS20130396 . [DOI] [PubMed] [Google Scholar]
- 46.Benton MC, Johnstone A, Eccles D, Harmon B, Hayes MT, Lea RA, et al. An analysis of DNA methylation in human adipose tissue reveals differential modification of obesity genes before and after gastric bypass and weight loss. Genome Biol. 2015;16:8 Epub 2015/02/05. 10.1186/s13059-014-0569-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eap CB, Crettol S, Rougier JS, Schlapfer J, Sintra Grilo L, Deglon JJ, et al. Stereoselective block of hERG channel by (S)-methadone and QT interval prolongation in CYP2B6 slow metabolizers. Clin Pharmacol Ther. 2007;81(5):719–28. Epub 2007/03/03. 10.1038/sj.clpt.6100120 . [DOI] [PubMed] [Google Scholar]
- 48.Kraev A, Quednau BD, Leach S, Li XF, Dong H, Winkfein R, et al. Molecular cloning of a third member of the potassium-dependent sodium-calcium exchanger gene family, NCKX3. J Biol Chem. 2001;276(25):23161–72. Epub 2001/04/11. 10.1074/jbc.M102314200 . [DOI] [PubMed] [Google Scholar]
- 49.Naour N, Rouault C, Fellahi S, Lavoie ME, Poitou C, Keophiphath M, et al. Cathepsins in human obesity: changes in energy balance predominantly affect cathepsin s in adipose tissue and in circulation. J Clin Endocrinol Metab. 2010;95(4):1861–8. Epub 2010/02/19. 10.1210/jc.2009-1894 . [DOI] [PubMed] [Google Scholar]
- 50.Araujo TF, Cordeiro AV, Vasconcelos DAA, Vitzel KF, Silva VRR. The role of cathepsin B in autophagy during obesity: A systematic review. Life Sci. 2018;209:274–81. Epub 2018/08/15. 10.1016/j.lfs.2018.08.024 . [DOI] [PubMed] [Google Scholar]
- 51.MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017;45(D1):D896–D901. Epub 2016/12/03. 10.1093/nar/gkw1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahashi N, Yoshizaki T, Hiranaka N, Kumano O, Suzuki T, Akanuma M, et al. The production of coagulation factor VII by adipocytes is enhanced by tumor necrosis factor-alpha or isoproterenol. Int J Obes (Lond). 2015;39(5):747–54. Epub 2014/12/17. 10.1038/ijo.2014.208 . [DOI] [PubMed] [Google Scholar]