Fig 4. Conserved structural interactions in EIAV and HIV-1.
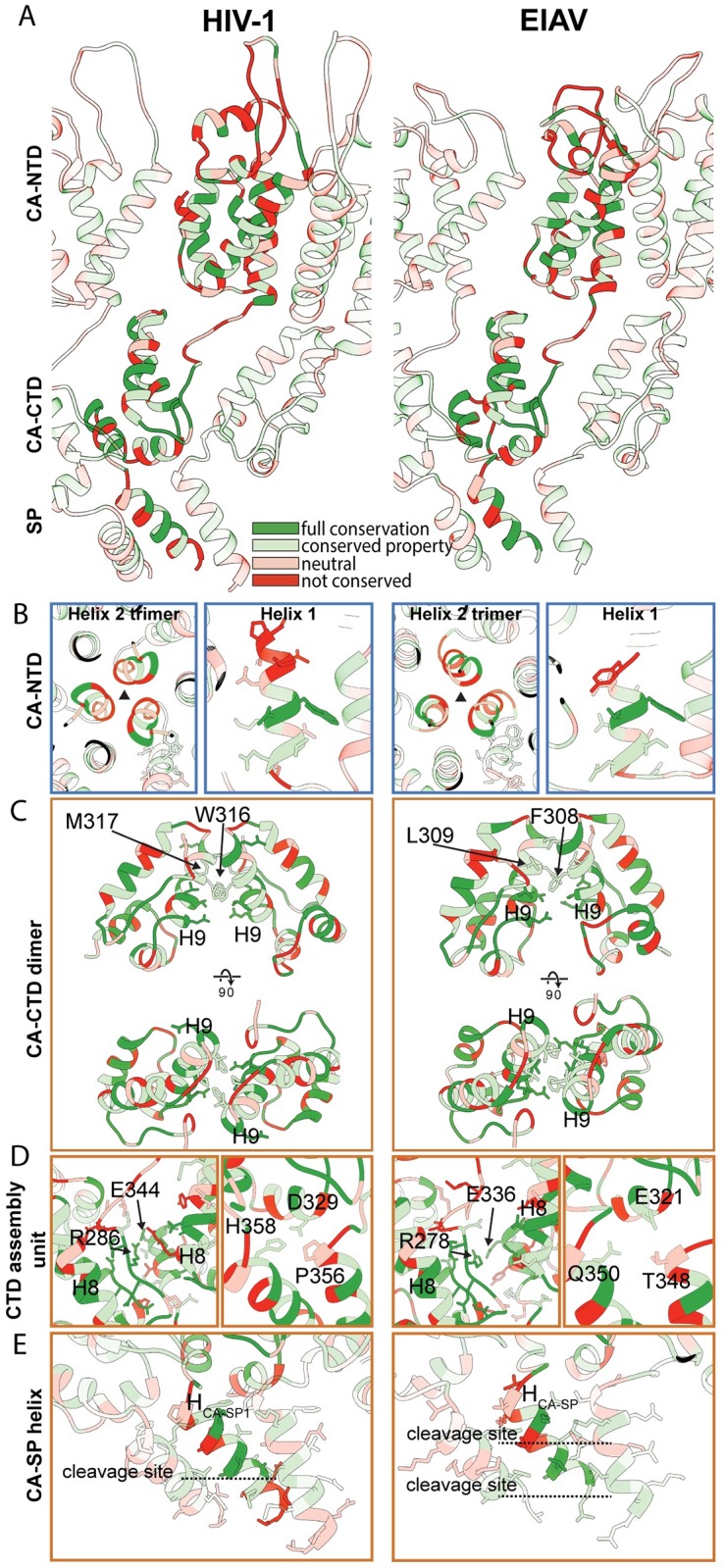
Comparison of structural features in EIAV and HIV-1. The HIV-1 CA-SP1 model derived from HIV-1 GagΔ16–99Δp6 (pdb 5l93, referred to as ΔMACANCΔp6 in [15]) is shown on the left. The EIAV CA-SP model derived from EIAV GagΔMA is shown on the right. S3 Movie shows a guided tour of this comparison. (A) Side view on the CASP lattice of EIAV and HIV-1 CASP. One monomer is highlighted, surrounding monomers of the lattice are shown with reduced opacity. The residues are colored according to the conservation between the two viruses. The color legend is indicated in panel (A). (B) Interactions and structural features in the CANTD. The trimeric interface in EIAV and HIV-1 is similar (Left). The Helix 1 in EIAV is shorter than in HIV-1 (Right). The extent of helix 1 in EIAV approximately corresponds to the conserved residues in HIV-1. (C) Comparison of the dimeric CACTD interface. Both lentiviruses use hydrophobic residues in helix 9 to stabilize the dimeric interface. F308/L309 and W316/M317 are annotated in EIAV and HIV-1, respectively. (D) Conserved residues in the MHR and helix 11 contribute to interactions around the hexameric ring to stabilize the immature CA assembly (Left). In HIV-1 residues D329, P356 and H358 form an important three-way interaction linking the CACTD base and the CA-SP1 helix of two adjacent CA monomers to each other. The equivalent residues in EIAV are E321, T348 and Q350 (Right). (E) The EIAV CASP 6HB is shorter than its counterpart in HIV-1. In EIAV and HIV-1 the CA-SP cleavage site is located within the helix, while the SP-NC cleavage site is located below the helix. Proteolytic cleavage sites are annotated by dashed lines.
