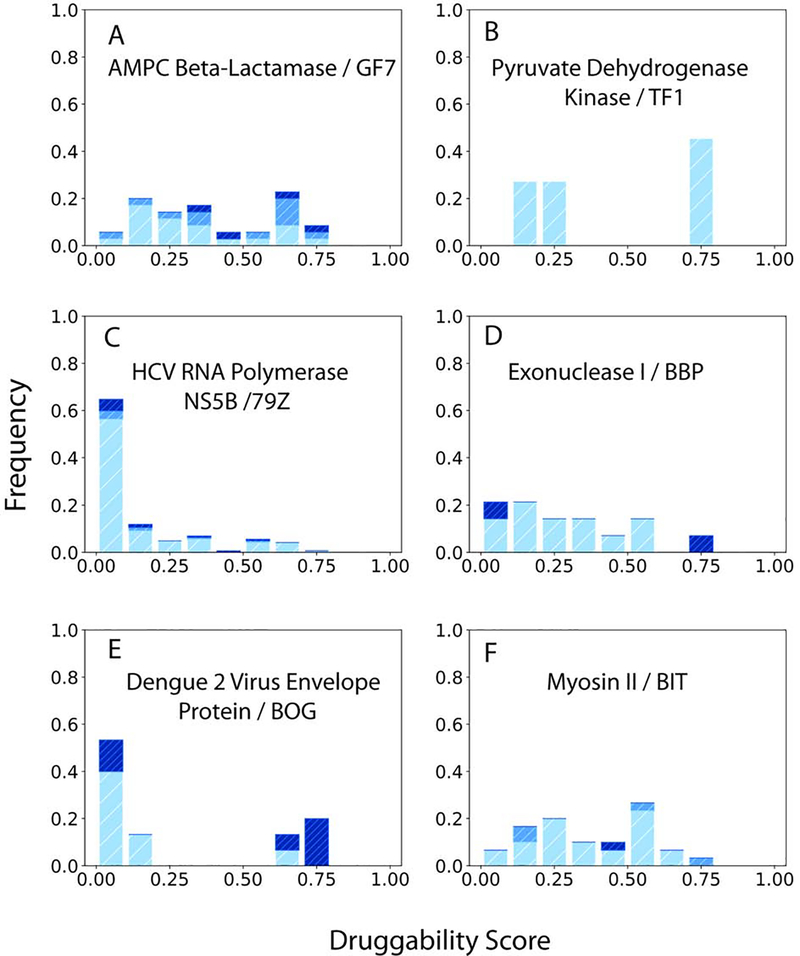
Figure 8.
Druggability scores (DSs) of unliganded structures of proteins with cryptic sites impacted by mutations or binding at distant sites. The ligand bound at the cryptic site shown in parenthesis. The distributions of DS values are shown in dark, light, and medium blue, respectively, for unbound structures, complexes, and mutants. A. AMPc beta-lactamase (Inhibitor GF7). B. Human pyruvate dehydrogenase kinase (Allosteric inhibitor TF1). C. Hepatitis C virus RNA polymerase NS5B (Inhibitor 79Z binding near the active site). D. Exodeoxyribonuclease I (Inhibitor BCBP). E. Dengue 2 virus envelope protein (Detergent n-octyl--D-glucoside). F. Myosin II (inhibitor blebbistatin).