Figure 1.
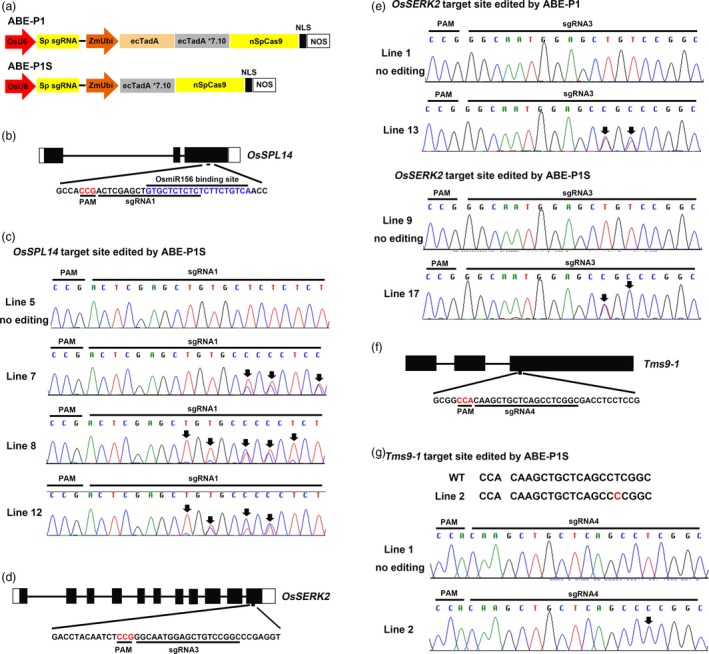
Adenine base editing at different target sites by ABE‐P1S in rice. a, Diagram of the ABE‐P1 and ABE‐P1S base editors used in this study. NLS, nuclear localization signal from Argobacterium VirD2 protein. b, Schematic view of the sgRNA1 target site in OsSPL14. c, Sequence chromatograms of Line 7, Line 8 and Line 12 at the OsSPL14 target site edited by ABE‐P1S. d, Schematic view of the sgRNA3 target site in OsSERK2. e, Sequence chromatograms of Lines 13 and 17 at the OsSERK2 target site edited by ABE‐P1 and ABE‐P1S, respectively. f, Schematic view of the sgRNA4 target site in Tms9‐1. g, One representative line, Line 2, edited by ABE‐P1S is a homozygous mutant. Sequence chromatogram of Line 2 at the target site in Tms9‐1 is shown here. The PAM sequence and miRNA binding site are highlighted in red and blue, respectively (b, d, f). Sequence chromatograms of lines with no mutations at the corresponding target sites are used as controls (c, e, g). Arrows point to the positions with an edited base (c, e, g).
