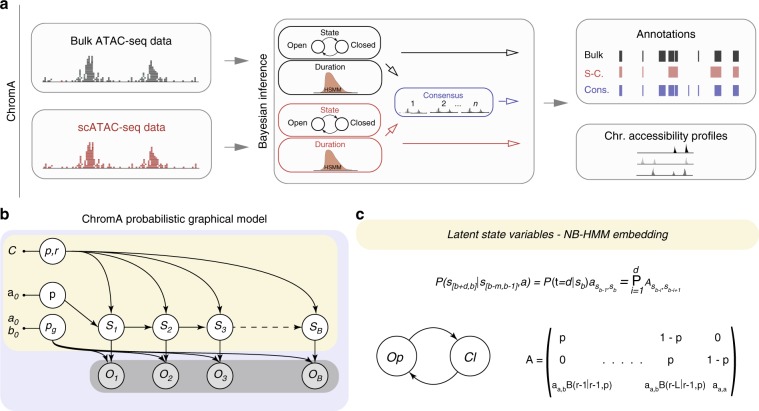
Fig. 1. Overview of chromatin accessibility annotation algorithm.
a ChromA is an easy to use algorithm that combines single and multiple BAM files (raw reads) or TSV files (list of Tn5-binding events) to create chromatin accessibility annotations. b Probabilistic graphical model describing ChromA’s structure. In this representation, nodes describe random variables and arrows depict dependencies among the variables. ChromA models the number of Tn5-binding events observed at each base using observed variables O (representing the number of binding event), and latent variables S (representing chromatin state). Subscripts denote base position, ranging from 1 to the length of a chromosome, B. Observed variables O are modeled using a geometric distribution with parameter pg. Chromatin-state variables S are subjected to semi-Markovian dynamics, depending on the previous chromatin state. π described the initial chromatin state, and p and r characterize the semi-Markovian transition matrix dictating chromatin-state context. c Our ATAC-seq pipeline using bulk measurements annotates chromatin using two states, open Op or closed Cl. Both states are characterized by semi-Markovian dynamics. The probability of annotating chromatin in bases b to b + d given previous chromatin states depends on two factors: the probability of transitioning between states, symbolized by transition matrix a, and the probability of dwelling in the new states during d bases. When the duration is characterized by a NB distribution with parameters p and r, the transition matrix can be re-written using an embedding matrix A. In the figure, we reproduce a simple transition matrix A, in which p and r are the NB parameters, a is the transition matrix between states, and B represents the binomial coefficient.