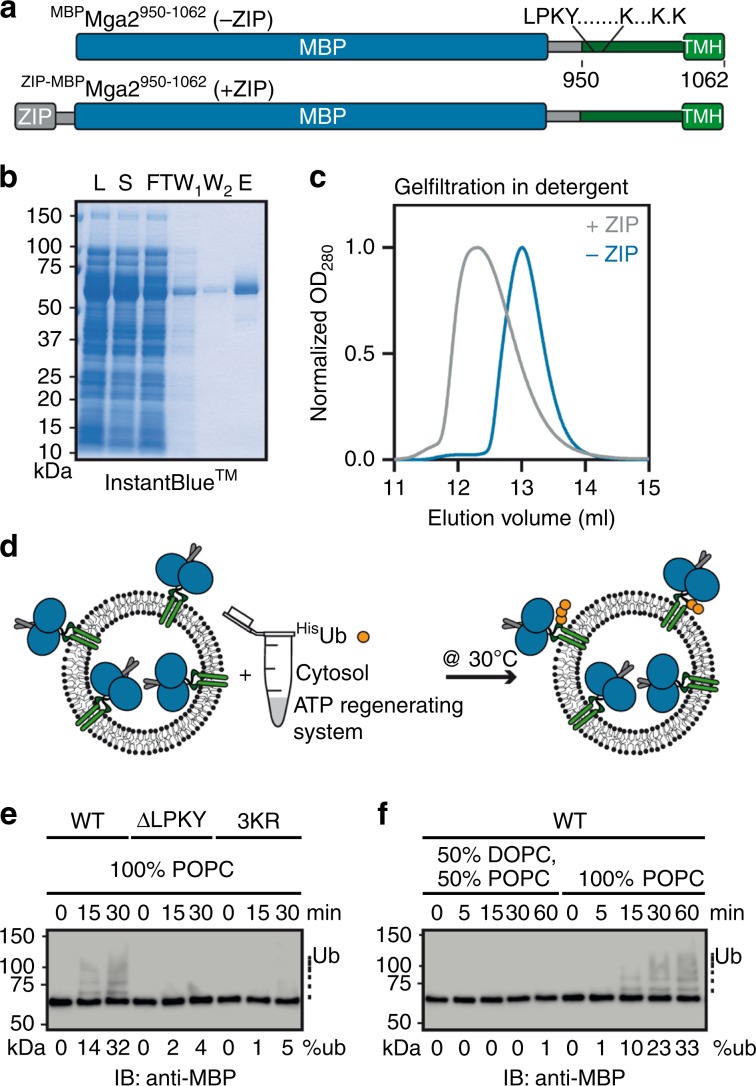
Fig. 2. An in vitro sense-and-response system for membrane lipid saturation.
a Schematic representation of the sense-and-response constructs. The fusion proteins are composed of the maltose-binding protein (MBP, blue) and Mga2950–1062 (green) with the Rsp5-binding site (LPKY), three lysine residues as targets of ubiquitylation (K980, K983, and K985), a predicted disordered juxtamembrane region, and the C-terminal TMH. An optional N-terminal leucine zipper derived from Gcn4 (gray, Gcn4249–281) supports dimerization. b Isolation of the zipped sense-and-response construct by affinity purification. 0.1 OD units of the lysate (L), soluble (S), flow-through (FT), and two wash fractions (W1,2), as well as 1 µg of the eluate were subjected to SDS-PAGE followed by InstantBlueTM staining. The protein was further purified by preparative SEC (Supplementary Fig. 1a). c One hundred micrograms in 100 µl of the purified sense-and-response constructs either with (+ZIP) or without zipper (−ZIP) were loaded onto a Superdex 200 10/300 Increase column (void volume 8.8 ml). d Schematic representation of the in vitro ubiquitylation assay. Proteoliposomes containing ZIP-MBPMga2950–1062 were mixed with 8xHisUbiquitin (HisUb), an ATP-regenerating system, and cytosol prepared from wild-type yeasts to facilitate Mga2 ubiquitylation at 30 °C. e The reaction was performed with the ZIP-MBPMga2950–1062 wild-type (WT) construct, a variant lacking the Rsp5-binding site (∆LPKY), and a variant with arginine residues instead of the lysine residues K980, K983, and K985 (3KR), thus lacking the target residues of ubiquitylation. These variants were reconstituted in liposomes composed of 100 mol% POPC at protein-to-lipid ratio of 1:5000. After the indicated times, the reactions were stopped using sample buffer and were subjected to SDS-PAGE. For analysis, an immunoblot using anti-MBP antibodies was performed. f Ubiquitylation reactions were performed as in e with the WT sense-and-response construct reconstituted in the indicated lipid environments at a molar protein-to-lipid ratio of 1:5000. Source data are provided as a Source Data file.