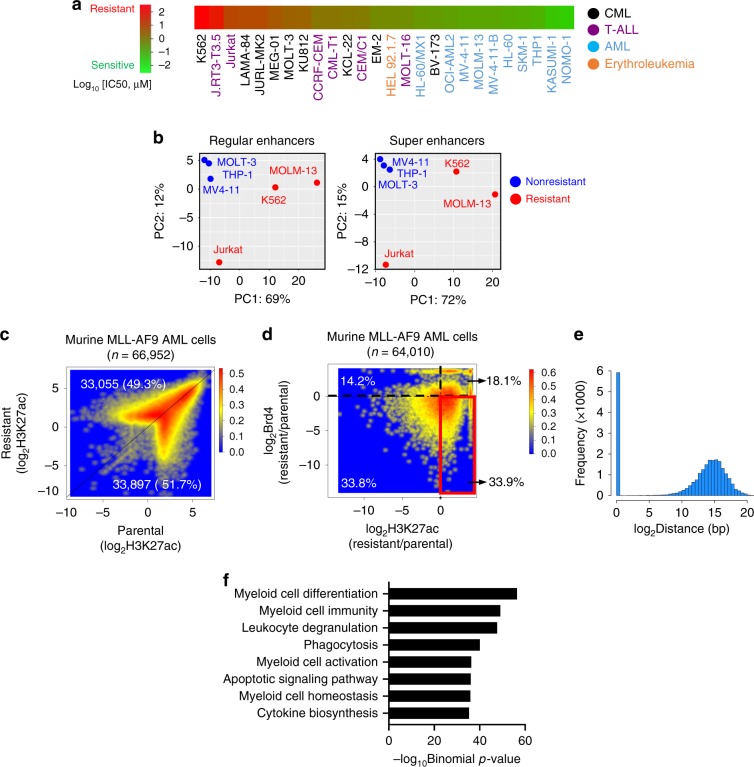
Fig. 1. BETi-resistant leukemia cells undergo BRD4-independent enhancer remodeling.
a Heatmap representing the IC50 values of the indicated leukemia cell lines to a BRD4 inhibitor JQ1. The IC50 values were obtained from the published database PHARMACODB. b The principal component analysis (PCA) of enhancer region distributions in the indicated BETi-sensitive and BETi-resistant leukemia cells. c Smoothed scatter plot representation of H3K27ac-enriched regions in paired BETi-sensitive (parental, x-axis) and BETi-resistant (y-axis) murine AF9 AML cells. Color code represents the dot density. d Smoothed scatter plot representation of differential H3K27ac-enriched regions and Brd4 enriched regions in paired BETi-sensitive (parental) and BETi-resistant murine AF9 AML cells. Red highlighted region: genomic regions displayed increased H3K27ac enrichment with no change or decreased Brd4 binding in BETi-resistant murine AF9 AML cells. All the publicly available H3K27ac and BRD4 ChIP-seq datasets used in Fig. 1b–d were summarized in Supplementary Table 1. Color code represents the dot density. e The histogram showing the distribution of distance between the genomic regions highlighted within the red box in Fig. 1d and their closest genes. f Genomic Regions Enrichment of Annotations Tool (GREAT) analysis of genomic regions highlighted in the red box of Fig. 1d.