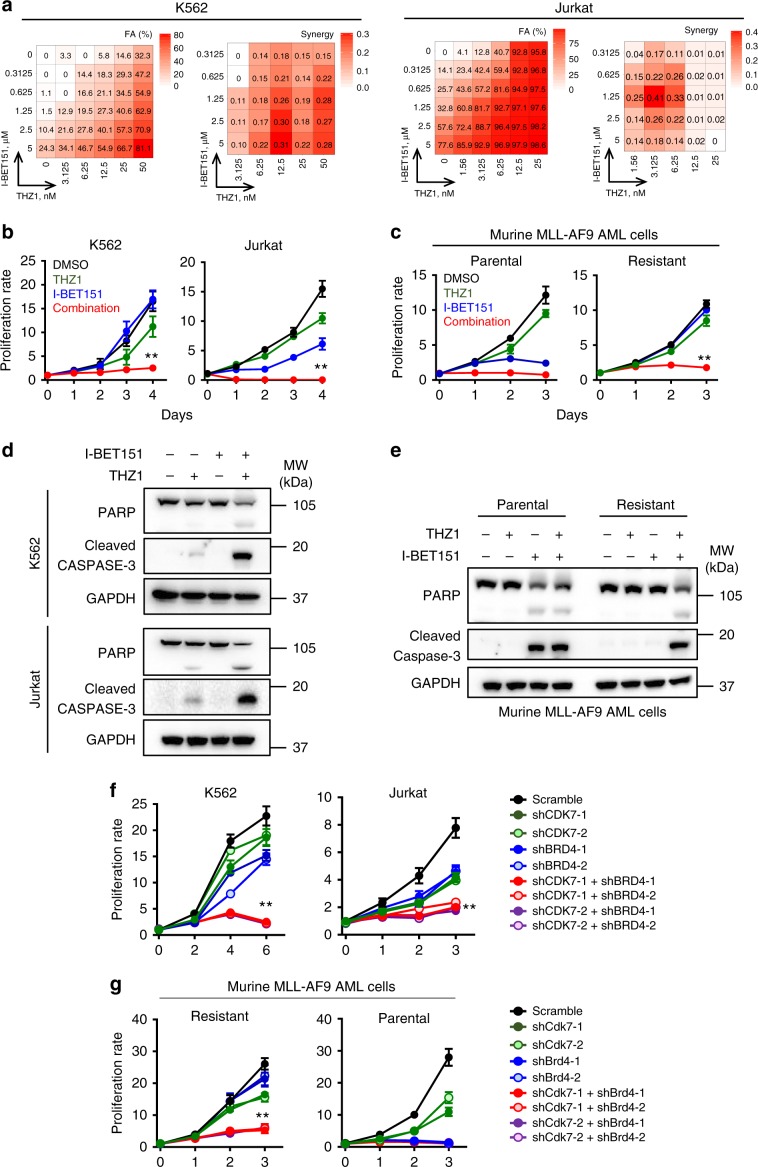
Fig. 2. Dual inhibition of CDK7 and BRD4 synergistically suppresses the growth of BETi-resistant leukemia cells.
a Heatmap representation of Fraction Affected (FA) and Bliss interaction index across five-point dose range of a BET inhibitor (I-BET151) and a CDK7 inhibitor (THZ1) in K562 and Jurkat cells. Mean values of triple biological experiments were shown. (b and c) Proliferation analysis of K562 and Jurkat cells (b) or murine AF9 AML cells (c) treated with DMSO (black), THZ1 (green), I-BET151 (blue), and the combination of THZ1 + I-BET151 (red). The concentrations of I-BET151 and THZ1 were 2.5 μM and 12.5 nM for K562, 2.5 μM and 3.125 nM for Jurkat, 2.5 μM and 50 nM for murine AF9 AML cells, respectively. Data were shown as mean ± S.D; n = 6 from 3 independent assays, **P = 0.00002 (K562), 0.00009 (Jurkat) and 0.000006 (AF9 resistant), by two-tailed Student’s t test. d, e Immunoblot analysis on apoptosis-related marker PARP and cleaved caspase 3 (C/Caspase3) in K562 (d, top), Jurkat (d, bottom) and murine AF9 AML cells (e) treated with DMSO, THZ1, I-BET151, and the combination of THZ1 + I-BET151. The inhibitor concentrations were the same as shown in Fig. 2b, c. Three independent assays were performed. f, g Quantification of proliferation of K562, Jurkat cells (f) and murine AF9 AML cells (g) transduced with shRNAs targeting CDK7 and/or BRD4. Data were shown as mean ± S.D; n = 8 from three independent assays, **P = 0.00003 (K562), 0.00001 (Jurkat) and 0.000002 (AF9 resistant), by two-tailed Student’s t test.