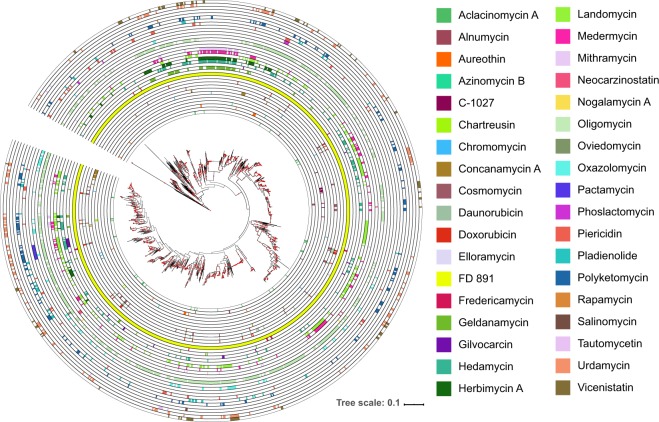
Figure 3.
Phylogenetic distribution of the CGCs that encode antitumor compounds. Presence of each CGC was determined by searching all the genomes for homologs of each of the genes comprising the CGCs using BLASTX24 with a minimum e-value of 10−10. Sequences of individual genes in a CGC were obtained from DoBISCUIT57 and were used as query sequences. Presence of the CGC was inferred if there were significant BLASTX hits for at least 90% of the individual genes within the CGC. The phylogenetic tree is identical to that used in Fig. 1.