Abstract
Background
Acute diarrhea is still a common and serious disease. The causes of acute diarrhea are very complicated. Therefore, we need to find a medicine to control diarrhea symptoms, save time for diagnosis of pathogens, and prevent drug abuse. Ellagic acid (EA), a natural polyphenol drug, has anti-diarrhea effects. However, the action mechanisms of EA for non-specific diarrhea have not been characterized.
Materials and Methods
To study the mechanisms of EA, mice were divided into four groups. Group C were intraperitoneally injected with 0.1 ml physiological saline and orally given 0.2 ml physiological saline, and then after experiment began 0.5 h, orally administered 0.3 ml physiological saline. Group D were intraperitoneally injected with 0.1 ml physiological saline and orally given 0.2 ml castor oil, and then after experiment began 0.5 h, orally administered 0.3 ml physiological saline. Group E were intraperitoneally injected with 0.1 ml physiological saline and orally given 0.2 ml castor oil, and then after experiment began 0.5 h, orally administered 0.3 ml EA (10 mg/ml). Group V were intraperitoneally injected with 0.1ml GW9662 (1m g/ml) and orally given 0.2 ml castor oil, and then after experiment began 0.5 h, orally administered 0.3 ml EA (10 mg/ml). Transcriptome were performed on ileum tissues of mice in group D and E. Histological examination and qRT-PCR were performed on ileum tissues of mice in group C, D, E, and V.
Results
We found that a total of 273 differentially expressed genes (DEGs) were obtained, including 160 up-regulated DEGs and 113 down-regulated DEGs. The DEGs were enriched in 458 Gene Ontology (GO) terms and 15 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, respectively. The peroxisome proliferator activated receptor (PPAR) signaling pathway was the most significantly enriched in KEGG pathways. We used the PPAR-specific antagonist GW9662 to validate the anti-diarrhea and anti-inflammatory effect of EA in group V compared with group E. Conclusively, EA protected ileums against castor oil-induced inflammation and diarrhea by activating the PPAR signaling pathway and a method was used to study the mechanism of EA.
Keywords: ellagic acid, transcriptome, PPAR signaling pathway, castor oil, diarrhea
Introduction
Diarrhea is the passage of liquid or loose stools more frequently than normal for an individual (Vrese and Offick, 2010). Acute diarrhea is a common and serious disease worldwide. Miller et al. (2002) reported that nosocomial clostridium difficile-associated diarrhea is a nosocomial infectious complication, has relationship with substantial morbidity and mortality, and causes a financial burden on healthcare institutions in Canada (Miller et al., 2002). In USA, diarrhea is an important cause of morbidity in this insured population of young children (Zimmerman et al., 2001). Curcio reviewed resent studies and found that there is a high incidence of clostridium difficile-associated diarrhea in developing countries (Africa–Middle East, developing Asia, Latin America, and China) (Curcio et al., 2019).
The causes of acute diarrhea are very complicated. Toxicosis, endocrine dyscrasia, viruses, bacteria, parasite, and medications can cause acute diarrhea. Acute infectious diarrhea is one of the most common diseases in pediatric age with relevant burden both in high- and in low-income countries (Lo Vecchio et al., 2019). Studies have reported that acute diarrhea was accompanied by an increase in inflammatory responses (Mazzolin et al., 2013; Dong et al., 2019) and oxidative stress (Jabri et al., 2016a). More importantly, castor oil-induced intestinal hypersecretion had a physiological response similar to acute diarrhea in the intestine (Mazzolin et al., 2013; Jabri et al., 2016a; Jabri et al., 2016b). Models used in studying diarrhea often employ castor oil in mice (Robert and Rao, 1996) or rats (Amabeoku and Bamuamba, 2010). Symptoms of castor oil-induced diarrhea are non-specific and these models have been widely used in the screening of natural anti-diarrheal drugs.
To treat acute diarrhea, the first thing is to find the causes. Therefore, screening non-specific anti-diarrhea drugs from natural plants will save time for finding diarrhea causes and targeted medicines. For example, Pandey et al. (2017) reported that 50% ethanolic extracts of grilled and dried fruits effectively inhibited diarrhea (Pandey et al., 2017). Due to increasing interest in plant-based traditional medicines, many researches are interested in exploring their activities and mechanisms of action (Zhao et al., 2018). Natural products boast many advantages, such as low cost, ease of availability, the ability to circumvent drug resistance, and relief of diarrhea symptoms. These advantages enable earlier pathogen diagnosis and treatment. For instance, many natural antidiarrheal plants contained ellagic acid (EA) (Singh et al., 2017). EA is the dilactone of hexahydroxydiphenic acid and a natural phenol antioxidant found in fruits and vegetables. EA has potential anti-inflammatory and antioxidant properties (Priyadarsini et al., 2002; Yim et al., 2016; Verotta et al., 2018) and has been shown to have a preventive effect against many diseases, such as neurodegenerative diseases, cancer and diarrhea in vitro and in vivo (Buniatian, 2003; Kannan and Quine, 2012; Ahmed et al., 2016).
Peroxisome proliferator-activated receptors (PPAR) belong to the nuclear receptor superfamily of ligand-inducible transcription factors. PPARs are classified into three subtypes α, β/δ, and γ. Studies showed that the biological functions of PPARs are complex. Yaribeygi et al. (2018) reported that activated PPAR-α restored anti-oxidant defense systems and improved diabetes-induced oxidative stress (Yaribeygi et al., 2018). Intriguingly, Zeinali et al. (2017) suggested that chrysin (CH), a plant polyphenolic compound, which acts as an agonist of PPAR-γ, can be used as an anti-inflammatory and anti-oxidative agent in immunopathological and physicochemical injuries (Zeinali et al., 2017). Cadmium caused high levels of inducible nitric oxide synthase (iNOS) activity, nitric oxide (NO) content, and apoptosis via PPAR-γ/PI3K/Akt pathway in chicken pancreas (Jin. et al., 2018). Compared to dinitrobenzene sulfonic acid (DNBS)-treated PPAR-α wild-type (WT) mice, DNBS-treated PPAR-α knockout mice (PPAR-αKO) mice experienced more hemorrhagic diarrhea, more weight loss, higher rate of the extent and severity of the histological signs of colon injury (Cuzzocrea et al., 2004). Sarnelli et al. (2018) also found that palmytoilethanolamide, via PPAR-α-dependent mechanism, resulted in a significant antidiarrheal activity in WT rats (Sarnelli et al., 2018). However, the effect of EA on the PPAR signaling pathway remains unclear.
Transcriptome analysis is a recently developed deep sequencing technique and involves an informatics approach to solve an experimental limitation (Martin and Wang, 2011). Transcriptome has been developed for many research fields, such as toxicology (Chen et al., 2018; Chen et al., 2019); Li et al. (2018) and Skaria et al. (2019) reported that transcriptome was used to investigated molecular mechanism of drugs (Li et al., 2018; Skaria et al., 2019). The purpose of this study was to evaluate the protective mechanism of EA against diarrhea by transcriptome analysis in mouse models. Furthermore, histopathological examination and redox biomarkers were determined to study the anti-inflammatory and antioxidative effects of EA. We used the PPAR-specific antagonist GW9662 to validate the anti-inflammatory effects of EA. Transcriptome and qRT-PCR results showed that the PPAR signaling pathway was involved in the preventive mechanism of EA against castor oil-induced diarrhea.
Materials and Methods
Animals and Treatments
BALB/c mice were purchased from Harbin Medical University (Harbin, China) and raised in the experimental animal facility of Northeast Agricultural University. All experimental processes about animals complied with EU Directive (2010/63/EU) and were approved by the Ethics Committee of Northeast Agricultural University of China (Protocol number: SRM-06). After being acclimated for two weeks, twenty-one days old healthy mice (20.1 ± 0.5 g) were randomly divided into four groups (ten mice each group). The control group (group C) were intraperitoneally injected with 0.1ml physiological saline and orally given 0.2 ml physiological saline, and then after experiment began 0.5 h, orally administered 0.3 ml physiological saline. The diarrhea group (group D) were intraperitoneally injected with 0.1ml physiological saline and orally given 0.2 ml castor oil, and then after experiment began 0.5 h, orally administered 0.3 ml physiological saline. The EA group (group E) were intraperitoneally injected with 0.1ml physiological saline and orally given 0.2 ml castor oil, and then after experiment began 0.5 h, orally administered 0.3 ml EA (10 mg/ml). The verification group (group V) were intraperitoneally injected with 0.1ml GW9662 (1 mg/ml) and orally given 0.2 ml castor oil, and then after experiment began 0.5 h, orally administered 0.3 ml EA (10 mg/ml).
Castor oil can cause frequent stooling within 4 h. Each group mice were placed in a beaker (5000 ml) with a weighed filter paper at the bottom for observation. Filter paper in each beaker was also changed at the same time and weighed to obtain the stool mass. The doses and pretreatment times were obtained from preliminary studies in our laboratory. All experiments were carried out once a day for 3 consecutive days in a quiet laboratory and the ambient temperature was 20.5 ± 1°C. At the third day, we sacrificed the mice for experiments. Ileum samples were collected from each group and washed with physiological saline solution (0.9% NaCl) on ice-cold plates. Samples from group D and E were prepared for mRNA sequencing. All the mouse ileums of four groups were collected for morphological examination, oxidative stress biomarkers and proinflammatory factor kit and qRT-PCR.
mRNA Sequencing and Analysis
Total RNA was extracted with TruSeq Stranded Total RNA Library Prep Kit (Illumina, USA). RNA concentration and total quantity were detected by Invitrogen Qubit 3.0 Spectrophotometer (Thermo Fisher Scientific, USA); RNA purity was checked using the Nanodrop 2000 (Thermo Fisher Scientific, USA); and RNA integrity was checked by Agilent 2100 Bioanalyzer (Agilent Technologies, USA). mRNA was obtained through purifying total RNA, and was broken into fragments of 100–300 bp. Reverse Transcribe was performed to synthesize First Strand cDNA using SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific, USA). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Then addition of adenlylate 3′ ends, connection sequencing adapters, selection library size, PCR amplification and checking library quality were performed in turn. Sequencing was performed on the Illumina Hiseq 2500 (Illumina, USA) and raw data was obtained. Raw data were first processed through in-house Perl scripts. Clean data were obtained by removing reads containing adapter sequences, poly-N sequences, and low-quality reads from raw data. The Q20, Q30, and GC content of the clean data were also calculated.
Reference genome and gene model annotation files were downloaded from the GenBank directly. An index of the reference genome was built using Bowtie v2.2.3. Paired-end clean reads were aligned using TopHat v2.0.12. Differential expression analysis of the two groups was performed using the DESeq R package (1.18.0). DESeq provided statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. Resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value < 0.05 found by DESeq were assigned as differentially expressed. GO and KEGG enrichment analysis of differentially expressed genes (DEGs). Gene Ontology (GO) enrichment analysis of differentially expressed genes (DEGs) was implemented by the GOseq R package, in which gene length bias was corrected. GO terms with corrected P-value less than 0.05 were considered significantly enriched by DEGs. KEGG was a database for understanding high-level functions and utilities of the biological system, such as the cell, the organism and the ecosystem, from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies (http://www.genome.jp/kegg/). We used KOBAS software to test the statistical enrichment of DEGs in KEGG pathways.
Morphological Examination in the Ileum
Ileum samples were cut into 0.5 cm × 0.5 cm tissue blocks, fixed in 10% formaldehyde, and embedded in paraffin. Sections were then stained with hematoxylin and eosin. Tissue slices were observed under a microscope by a pathologist blinded to the experiment (Hu et al., 2018).
Redox Biomarker Determinations
Ileum samples were homogenized in physiological saline, centrifuged at 3,000 g for 15 min, and supernatants were collected. The activities of superoxide dismutase (SOD) and glutathione peroxidase (GPx) and the content of malondialdehyde (MDA) were determined. Commercial assay kits for SOD (Superoxide Dismutase assay kit (A001-1)), GPx (Glutathione Peroxidase (GPX) assay kit (A005)) and MDA (Malondialdehyde (MDA) assay kit (A003-1)) were provided by Jiancheng Biotechnology Research Institute (Nanjing, China) (Su et al., 2018).
qRT-PCR Analysis
To validate the reliability of the RNASeq results, we selected Ccr6, Cd36, Cyp2e1, GPx, H2-Ob, interleukin (IL)-1β, IL-6, NF-κB, PPAR-γ, Sod (SOD), and TNFα genes for qRT-PCR. A housekeeping gene (β-actin) was used as a reference. Primer information for qRT-PCR are shown in Table 1 . Reactions were incubated in a Light Cycler® 480 System (Roche, Basel, Switzerland). Reactions contained 10 μl 2×SYBR Green I PCR Master Mix (Roche, Basel, Switzerland), 2 μl of diluted cDNA, 0.4 μl of each primer (10 μM), 0.4 μl 50×ROX reference Dye II and 6.8 μl PCR-grade water. PCR cycling conditions were: one cycle at 95 °C for 30 s, followed by 40 cycles at 95 °C for 15 s and 60 °C for 30 s. The amplification efficiency for each gene was determined using the DART-PCR program. Relative mRNA abundance was calculated according to the Pfaffl method (Pfaffl, 2001).
Table 1.
Gene-special primers used in qRT-PCR.
| Gene | Forward (5 → 3) | Reverse (5 → 3) |
|---|---|---|
| Ccr6 | GTGTGGCAGTGTGGTTCATCTCC | GTGGCTCACAGACATCACGATCC |
| Cd36 | GCGACATGATTAATGGCACAGACG | CCGAACACAGCGTAGATAGACCTG |
| Cyp2e1 | AAGGACGTGCGGAGGTTTTCC | TACATGGGTTCTTGGCTGTGT |
| GPx | CGCTTTCGTACCATCGACATC | GGGCCGCCTTAGGAGTTG |
| H2-Ob | CACAACCTGCTGCTCTGCTCTG | GACCTCTCCTCCTGTCCATTCCG |
| IL-1β | GCAACTGTTCCTGAACTCAACT | ATCTTTTGGGGTCCGTCAACT |
| IL-6 | GGAGCCCACCAAGAACGATA | ACCAGCATCAGTCCCAAGAA |
| NF-κB | TCTCTATGACCTGGACGACTCTT | GCTCATACGGTTTCCCATTTAGT |
| PPAR-γ | CCAGAGCATGGTGCCTTCGCT | CAGCAACCATTGGGTCAGCTC |
| Sod | GTG ATTGGG ATTGCGCAG TA | TGGTTTGAG GGTAGCAGATGAGT |
| TNF-α | CCCTCACACTCAGATCATCTTCT | GCTACGACGTGGGCTACAG |
| β-actin | GTGCTATGTTGCTCTAGACTTCG | ATGCCACAGGATTCCATACC |
Statistical Analysis
Statistical analysis was performed using SPSS software (version 20.0, SPSS Inc., Chicago, IL, USA). Statistical significance was evaluated by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. Data are expressed as mean ± standard deviation (SD).
Results
Animal Observations
We used the indexes of mass of stool, fecal output, onset of diarrhea, number of animals exhibiting diarrhea, and percentage episode inhibition to evaluate diarrhea symptoms, as shown in Table 2 . No diarrhea symptoms were observed in mice of group C. Mass of stool were significantly decreased in group E (P < 0.01) and V (P < 0.05) compared with that in group D. EA effectively inhibits diarrhea onset of diarrhea in group E compared with that in group D (P < 0.01), while there was no difference in Onset of diarrhea between group D and group V (P > 0.05).
Table 2.
Effect of EA on castor oil-induced diarrhea in mice.
| Group | C | D | E | V |
|---|---|---|---|---|
| Mass of stool (Mean ± SD (g)) | 0 | 0.75 ± 0.12 | 0.23 ± 0.02** | 0.65 ± 0.03* |
| Fecal output (%) | 0 | 100 | 30.67 | 86.67 |
| Onset of diarrhea (Mean ± SD (min)) | – | 30.12 ± 1.96 | 84.74 ± 3.84** | 35.33 ± 6.20 |
| No. of animals exhibiting diarrhea | 0 | 10/10 | 2/10 | 10/10 |
| Percentage Episode inhibition (%) | – | 0 | 80.0 | 0 |
*represented significant difference (P < 0.05). **represented significant difference (P < 0.01). Statistical significance was evaluated by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test.
Morphological Structure
Morphological structure of group C, D, E, and V were shown in Figure 1 . The epithelial cell structures of ileums were arranged neatly and clear in group C ( Figure 1A ) and E ( Figure 1C ) compared with group D ( Figure 1B ) and V ( Figure 1D ). The numbers of goblet cells (GC) were increased and lots of lymphocytes (Ly) were aggregated in group D ( Figure 1B ) and V ( Figure 1D ). The tissues showed cellulose-like swelling (CLS), uneven staining, and inflammatory exudation in group D ( Figure 1B ) and V ( Figure 1D ).
Figure 1.
Ileums slices using HE staining. (A–D) were in the 400 × magnification.
Redox Biomarker Determination
We evaluated the systemic oxidative balance by redox biomarker determination in groups C, D, E and V. Levels of SOD, GPx, and MDA in ileum tissues of group C, D, E, and V mice were shown in Figure 2 . Castor oil induced oxidative damage of the ileums, while EA significantly alleviated castor oil-induced oxidative damage. SOD activity was significantly reduced in group D and V (P < 0.01). There were no differences between groups C and E. Compared to group C, castor oil significantly decreased GPx content in group D and V (P < 0.01), with no significant difference in group E (P > 0.05). Compared to group C, castor oil significantly increased MDA content in group D and V (P < 0.01).
Figure 2.
Determination of redox biomarker. **represented significant difference (P < 0.01). Statistical significance was evaluated by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test.
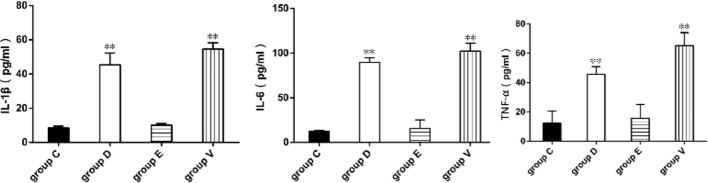
Proinflammatory Factor Determination
IL-1β, IL-6, and TNF-α followed similar trends in the four experimental groups, as shown in Figure 3 . IL-1β, IL-6, and TNF-α levels were higher in group V and D compared those with group C (P < 0.01), while IL-1β, IL-6 and TNF-α levels were no significantly increased in group E compared with that in group C (P > 0.05).
Figure 3.
Determination of proinflammatory factors. **represented significant difference (P < 0.01). Statistical significance was evaluated by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test.
mRNA Sequencing and Analysis
Libraries from the group D and E were mapped using TopHat2. The characteristics of these libraries are summarized in Table 3 . There were 106,567,752 and 100,936,974 raw reads and 116,141,362 and 104,686,394 raw reads were obtained from group D and E respectively. A total of 106,530,110 and 100,899,108 clean reads were obtained from group D and 116,125,974 and 104,665,316 clean reads were obtained from the group E. Clean base ratios were 99.96% in group D and 99.99 and 99.98% in group E. Parameters of Q20 were 97.51 and 97.12% in group D and 97.69 and 97.57% in group E and Q30 were 93.68, and 92.74% in group D and 94.21 and 93.95% in group E.
Table 3.
Summary statistics of the transcriptome sequencing from group D and group E.
| Sample | Raw reads | Clean reads | Clean base ratio (%) | Error rate(%) | Q20 (%) | Q30 (%) | GC content(%) |
|---|---|---|---|---|---|---|---|
| D1 | 106,567,752 | 106,530,110 | 99.96 | 0.0205 | 97.51 | 93.68 | 53.58 |
| D2 | 100,936,974 | 100,899,108 | 99.96 | 0.0356 | 97.12 | 92.74 | 53.71 |
| E1 | 116,141,362 | 116,125,974 | 99.99 | 0.0286 | 97.69 | 94.21 | 52.29 |
| E2 | 104,686,394 | 104,665,316 | 99.98 | 0.0217 | 97.57 | 93.95 | 52.44 |
Differential Expression Analysis
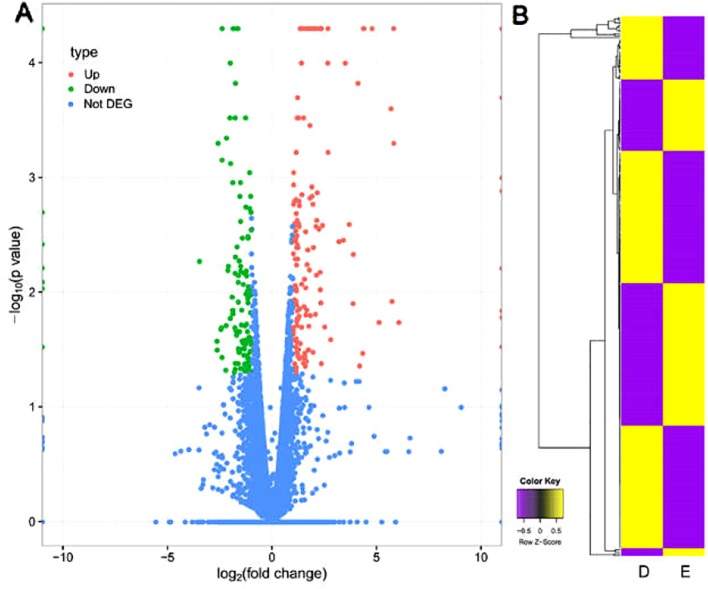
RNA-Seq reads mapped to mouse reference genome were aligned and their relative abundances were estimated using TopHat2. According to the statistical analysis of unigene data with differentially expressed genes sequencing, genes significantly and differentially expressed between group E and group D were identified. A fold-change in gene expression >2 and P < 0.05 were considered to be differential expressed genes (DEGs). The results revealed that 273 genes were differentially expressed, including 160 up-regulated and 113 down-regulated genes. The volcano plot in Figure 4A showed the distribution trends for DEGs (green spots represented down-regulated genes; red spots represented up-regulated genes) and non-DEGs (blue spots).
Figure 4.
The figure of DEGs. (A) Volcano plot of distribution trends for DEGs in group D and group E. The log2 [fold change (group D/group E)] indicated the mean expression level for each gene. Each dot represented one gene. Red dots represented upregulated genes and green dots represented down-regulated genes. Blue dots represented genes with no differential expression. (B) Hierarchal clustering heat map of group D and group E.
DEGs were analyzed by hierarchal clustering heatmap analysis, and genes with the same or similar expression behavior were clustered ( Figure 4B ). The hierarchal clustering heatmap were shown log2(transformed FPKM values) for all 273 DEGs in group D and group E. Each column represented group D or E and each row represented a DEG (yellow denoted upregulation; purple denoted downregulation; and the color scale was at the bottom). Expression values are mean-centered.
GO and KEGG Enrichment Analysis of DEGs
A total of 458 GO terms between groups D and E were significantly enriched with GO analysis, including 358 biological process (BP) terms, 51 cellular component (CC) terms, and 49 molecular function (MF) terms. We selected the top 10 GO terms from each of the BP, CC and MF subgroups based on significance (P-value), as shown in Figure 5A . For BP terms, the represented categories were immune system processes (GO: 0002376), regulation of immune system processes (GO: 0002682) and small molecule metabolic processes (GO: 0044281). For CC terms, the represented categories were extracellular region (GO: 0005576), extracellular region part (GO: 0044421) and extracellular organelle (GO: 0043230). For MF terms, the represented categories were oxidoreductase activity (GO: 0016491), hormone activity and carboxylic acid binding (GO: 0031406) ( Figure 5A ). A number of unigenes were also involved in binding, oxidoreductase activity and immune immunologic systems and anti-inflammation signaling, which suggested that these unigenes may play a role in EA protection and associated impacts. We selected the top 30 DEGs from each subgroups based on significance (P-value). Heatmaps of the top 30 DEGs were plotted using their log2-transformed FPKM values (Red represented upregulation; Blue represented downregulation in Figure 5B ).
Figure 5.
GO and KEGG analysis of DEGs. (A) The top 10 GO terms of BP, MF, and CC. (B) The top 30 DEGs heatmaps of BP, MF, and CC. (C) 15 pathways of DEGs using KEGG enrichment analysis.
Many of the DEGs were found to play a role in redox, immunity, lipid metabolism and inflammation. DEGs in mice from groups D and E were further annotated by KEGG, which found that 15 pathways were significant (P < 0.05). Figure 5C showed KEGG enrichment analysis of DEGs. Compared with whole genome expression, the PPAR signaling pathway was found to be the most significantly enriched pathway.
Validation of RNA-Seq Results with qRT-PCR
The 3 genes (Cyp2e1, SOD and GPx) were identified to be associated with oxidative stress. The 5 genes (CD36, NF-κB, IL-1β, IL-6, and TNF-α) were identified to be associated with inflammation. The 2 genes (H2-Ob and Ccr6) were identified to be associated with immune functions. Fold-changes in qRT-PCR were compared with RNA-Seq expression profiles. The log2 (fold-change) values of the genes identified by RNA-seq and qRT-PCR were: Ccr6 (2.24 vs. 1.41), CD36 (–2.34 vs. –0.64), Cyp2e1 (–1.43 vs. –0.67), GPx (1.28 vs. 1.03), H2-Ob (2.18 vs. 0.67), IL-1β (–1.25 vs. –0.54), IL-6 (–1.66 vs. –0.72), NF-κB (–1.64 vs. –0.72) PPAR-γ (2.76 vs. 1.87), SOD (1.57 vs. 1.06) and TNF-α (–0.12 vs. –0.18). As shown in Figure 6 , the qRT-PCR results were consistent with the high-throughput sequencing data, suggesting that the transcriptome sequencing data was reliable.
Figure 6.

Comparison of DEGs and qRT-PCR confirmation.
Discussion
Diarrhea is a disease which can cause death in humans (Emerson and Savage, 2017) and animals (Wolff et al., 2017). Transcriptome analysis was used to study the mechanism of action of anti-diarrheal drugs (Men et al., 2018; Wu et al., 2018; Yao et al., 2018), while the mechanism of action of EA in the treatment of diarrhea remains unclear. In this study, we used EA to protect small intestine against castor oil-induced diarrhea in a mouse model. Deep transcriptome sequencing was used to analyze the transcriptomic profiles of ileum tissues in diarrhea model with or without EA treatment. We identified 273 DEGs, including 160 up-regulated genes and 113 down-regulated genes. DEGs were annotated by GO in mice from group E compared with group D. A total of 458 GO terms between groups D and E were significantly enriched. According to P-value, the top 30 DEGs were selected from each sub-category (BP, CC, and MF). There were 15 pathways annotated by KEGG in mice from group E compared with group D. PPAR-γ was the most significant signaling pathway. In the study, we used transcriptome to study the mechanism of EA protecting ileum and inhibiting diarrhea.
Many hazardous materials induced oxidative stress that caused inflammation or injury (Wang et al., 2019; Qu et al., 2019). In our study, Castor oil induced oxidative stress, decreased SOD and GPx levels, and increased MDA levels in group D compared with group C and E. Celik et al. (2013) reported that EA decreased the activity of Cyp2e1 and increased the activity of GPx (Celik et al., 2013). Jabri et al. (2016b) reported similar results (Jabri et al., 2016b). SOD and GPx activities were increased in EA-treated V79-4 cells (Han et al., 2006). Expression of PPAR-α was increased, whereas that of Cyp2e1 was reduced (Nakamuta et al., 2005; Aristatile et al., 2014). Yuce et al. (2007) found that the administration of EA to cisplatin‐treated rats decreased the MDA levels, and increased GPx and CAT in liver and heart tissue of rats (Yuce et al., 2007). In our experiment, EA increased expressions of PPAR-γ, SOD, GPx and decreased MDA level and Cyp2e1 expression, which suggested that EA decreased oxidative stress by PPAR signaling pathway. Cd36 is a membrane glycoprotein, which presented on some epithelia and contributed to inflammatory responses (Silverstein and Febbraio, 2009). Cd36 was a key modulator of proinflammatory and oxidative pathways. Silverstein and Febbraio (2009) found that Cd36-deficient mice exhibited levels of activated NF-κB and oxidative stress decreased in chronic kidney disease (CKD) (Silverstein and Febbraio, 2009). In our experiment, EA decreased Cd36, NF-κB, IL-1β, IL-6, and TNF-α, which suggested that EA can inhibit inflammation, protect the small intestine and treat diarrhea. Ccr6 was expressed on immature dendritic cells and B cells and memory T cells and involved in mucosal immune responses (Puleston et al., 2005), and a common marker for Th1/Th17 cells, which also preferentially expressed the nuclear receptor PPAR-γ (Bernier et al., 2013). The non-classical major histocompatibility complex (MHC) class II gene H2-Ob was enriched in antigen processing/presentation pathways (Stables et al., 2011). Additionally, H2-Ob is thought to be a switching gene in innate immunity (Khayer et al., 2017). In our experiment, EA increased Ccr6 and H2-Ob expression. Ramirez et al. (2017) reported that tea made from Mangifera indica L. leaves of the Uba variety upregulated PPAR-γ, exerting antioxidant and anti-inflammatory effects (Ramirez et al., 2017). PPAR regulated the genes of Ccr6, Cd36, Cyp2e1, and H2-Ob. EA activated PPAR signaling pathway and treated diarrhea, by Inhibiting oxidative stress, inhibiting inflammation and improving immunity. Interestingly, GW9662, a PPAR antagonist, inhibited the effect of EA on diarrhea and inflammation in group V, which suggested that EA treatmented of diarrhea through PPAR signaling pathway. Yang et al. (2014) reported that some PPAR-γ ligands, such as emodin and protocatechuic acid, were used to attenuate inflammation by activating PPAR-γ, which was activated by the MAPK/NF-κB pathway (Yang et al., 2014). Furthermore, activated NF-κB mediated the expression of a number of rapid response genes, such as iNOS and COX-2, involved in the inflammatory response to injury (Shi et al., 2019; Hu et al., 2019);. The expressions of COX-2 and iNOS can increased the expression of IL-1β, IL-6, and TNF-α (Horie et al., 2009). Consistent with this notion, our result of RNA sequencing showed that EA increased the expressions of PPAR-γ and decreased the expressions of IL-1β, IL-6, NF-κB, and TNF-α, which were proved by qRT-PCR.
In conclusion, transcriptome analysis of the ileum in a mouse diarrhea model successfully identified a large number of DEGs. These genes showed that EA mainly treated diarrhea by activating the PPAR signaling pathway.
Data Availability Statement
The datasets generated for this study can be found in NCBI GEO accession GSE142063.
Ethics Statement
All experimental processes about animals complied with EU Directive (2010/63/EU) and were approved by the Ethics Committee of Northeast Agricultural University of China (Protocol number: SRM-06).
Author Contributions
JC did the experiment and wrote the manuscript. HY and ZS designed and funded this experiment.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31572559) and (Grant No. 31802227); Postdoctoral scientific research developmental fund of Heilongjiang Province in 2018 (LBH-Q18020).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge their Bachelor’s student, Zhenfei Yi, for preparing the experiments and wish him an admission to graduate school.
References
- Ahmed T., Setzer W. N., Nabavi S. F., Orhan I. E., Braidy N., Sobarzosanchez E., et al. (2016). Insights into effects of ellagic acid on the nervous system: a mini review. Curr. Pharm. Des. 22, 1350–1360. 10.2174/1381612822666160125114503 [DOI] [PubMed] [Google Scholar]
- Amabeoku G. J., Bamuamba K. (2010). Evaluation of the effects of olea europaea L. subsp. africana (Mill.) P.S. Green (Oleaceae) leaf methanol extract against castor oil-induced diarrhoea in mice. J. Pharm. Pharm. 62, 368–373. 10.1211/jpp.62.03.0012 [DOI] [PubMed] [Google Scholar]
- Aristatile B., Al-Assaf A. H., Pugalendi K. V. (2014). Carvacrol ameliorates the PPAR-A and cytochrome P450 expression on D-galactosamine induced hepatotoxicity rats. Afr. J. Tradit. Complement Altern Med. 11, 118–123. 10.4314/ajtcam.v11i3.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier A., Cleret-Buhot A., Zhang Y., Goulet J. P., Monteiro P., Gosselin A., et al. (2013). Transcriptional profiling reveals molecular signatures associated with HIV permissiveness in Th1Th17 cells and identifies peroxisome proliferator-activated receptor gamma as an intrinsic negative regulator of viral replication. Retrovirology 10, 160. 10.1186/1742-4690-10-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buniatian G. H. (2003). Stages of activation of hepatic stellate cells: effects of ellagic acid, an inhibiter of liver fibrosis, on their differentiation in culture. Cell Proliferation 36, 307–319. 10.1046/j.1365-2184.2003.00287.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik G., Semiz A., Karakurt S., Arslan S., Adali O., Sen A. (2013). A comparative study for the evaluation of two doses of ellagic acid on hepatic drug metabolizing and antioxidant enzymes in the rat. Biomed. Res. Int. 2013, 358945. 10.1155/2013/358945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Xu Y., Han Q., Yao Y., Xing H., Teng X. (2018). Immunosuppression, oxidative stress, and glycometabolism disorder caused by cadmium in common carp (Cyprinus carpio L.): Application of transcriptome analysis in risk assessment of environmental contaminant cadmium. J. Hazard Mater 366, 386–394. 10.1016/j.jhazmat.2018.12.014 [DOI] [PubMed] [Google Scholar]
- Chen M., Li X., Shi Q., Zhang Z., Xu. S. (2019). Hydrogen sulfide exposure triggers chicken trachea inflammatory injury through oxidative stress-mediated FOS/IL8 signaling. J. Hazard Mater 368, 243–254. 10.1016/j.jhazmat.2019.01.054 [DOI] [PubMed] [Google Scholar]
- Curcio D., Cane A., Fernandez F. A., Correa J. (2019). Clostridium difficile-associated Diarrhea in Developing Countries: A Systematic Review and Meta-Analysis. Infect. Dis. Ther. 8, 87–103. 10.1007/s40121-019-0231-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzzocrea S., Di Paola R., Mazzon E., Genovese T., Muia C., Centorrino T., et al. (2004). Role of endogenous and exogenous ligands for the peroxisome proliferators activated receptors alpha (PPAR-α) in the development of inflammatory bowel disease in mice. Lab. Invest. 84 (12), 1643. 10.1038/labinvest.3700185 [DOI] [PubMed] [Google Scholar]
- Dong N., Xu X., Xue C., Wang C., Shan A. (2019). Ethyl pyruvate inhibits LPS induced IPEC-J2 inflammation and apoptosis through p38 and ERK1/2 pathways. Cell Cycle. (Georgetown Tex.) 18 (5), 1–15. 10.1080/15384101.2019.1653106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson E., Savage A. (2017). Acute respiratory infection, diarrhoea and fever in young children at-risk of intellectual disability in 24 low- and middle-income countries. Public Health 142, 85–93. 10.1016/j.puhe.2016.10.014 [DOI] [PubMed] [Google Scholar]
- European Commission (2010). Directive 2010/63/EU of the European Parliament and of the council of 22 September 2010 and of the council of 22 September 2010 on the protection of animals used for scientific purposes. Available at: https://eur-lex.europa.eu/legal-content/EN/TXT/HTML/?uri=CELEX:32010L0063&qid=1579452547508&from=EN. [Google Scholar]
- Han D. H., Lee M. J., Kim J. H. (2006). Antioxidant and apoptosis-inducing activities of ellagic acid. Anticancer Res. 26, 3601–3606. [PubMed] [Google Scholar]
- Horie A., Nagai K., Ohkura S., Ohama T., Komatsu H., Sato K. (2009). Proinflammatory cytokines suppress the expression level of protease-activated receptor-2 through the induction of iNOS in rat colon. J. Vet. Med. Sci. 71, 1609–1615. 10.1292/jvms.001609 [DOI] [PubMed] [Google Scholar]
- Hu X., Chi Q., Wang D., Chi X., Teng X., Li S. (2018). a. Hydrogen sulfide inhalation-induced immune damage is involved in oxidative stress, inflammation, apoptosis and the Th1/Th2 imbalance in broiler bursa of fabricius. Ecotoxicol. Environ. Saf. 164, 201–209. 10.1016/j.ecoenv.2018.08.029 [DOI] [PubMed] [Google Scholar]
- Hu X., Chi Q., Liu Q., Wang D., Zhang Y., Li S. (2019). b. Atmospheric H2S triggers immune damage by activating the TLR-7/MyD88/NF-κB pathway and NLRP3 inflammasome in broiler thymus. Chemosphere 237, 124427. 10.1016/j.chemosphere.2019.124427 [DOI] [PubMed] [Google Scholar]
- Jabri M. A., Rtibi K., Ben-Said A., Aouadhi C., Hosni K., Sakly M., et al. (2016. a). Antidiarrhoeal, antimicrobial and antioxidant effects of myrtle berries (Myrtus communis L.) seeds extract. J. Pharm. Pharmacol. 68, 264–274. 10.1111/jphp.12505 [DOI] [PubMed] [Google Scholar]
- Jabri M. A., Rtibi K., Sakly M., Marzouki L., Sebai H. (2016. b). Role of gastrointestinal motility inhibition and antioxidant properties of myrtle berries (Myrtus communis L.) juice in diarrhea treatment. Biomed. Pharmacother. 84, 1937–1944. 10.1016/j.biopha.2016.11.008 [DOI] [PubMed] [Google Scholar]
- Jin. X., Jia T., Liu R., Xu S. (2018). The antagonistic effect of selenium on cadmium-induced apoptosis via PPAR-γ/PI3K/Akt pathway in chicken pancreas. J. Hazardous Mater. 357, 355–362. 10.1016/j.jhazmat.2018.06.003 [DOI] [PubMed] [Google Scholar]
- Kannan M. M., Quine S. D. (2012). Ellagic acid protects mitochondria from β-adrenergic agonist induced myocardial damage in rats; evidence from in vivo, in vitro and ultra structural study. Food Res. Int. 45, 1–8. 10.1016/j.foodres.2011.09.018 [DOI] [Google Scholar]
- Khayer N., Marashi S. A., Mirzaie M., Goshadrou F. (2017). Three-way interaction model to trace the mechanisms involved in Alzheimer’s disease transgenic mice. PLoS One 12, e0184697. 10.1371/journal.pone.0184697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Luo M., Hu M., Guo A. Y., Yang X., Zhang Q., et al. (2018). Investigating the molecular mechanism of aqueous extract of cyclocarya paliurus on ameliorating diabetes by transcriptome profiling. Front. Pharmacol. 9, 912. 10.3389/fphar.2018.00912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Vecchio A., Buccigrossi V., Fedele M. C., Guarino A. (2019). Acute infectious diarrhea. Adv. Exp. Med. Biol. 1125, 109–120. 10.1007/5584_2018_320 [DOI] [PubMed] [Google Scholar]
- Martin J. A., Wang Z. (2011). Next-generation transcriptome assembly. Nat. Rev. Genet. 10, 671–682. 10.1038/nrg3068 [DOI] [PubMed] [Google Scholar]
- Mazzolin L. P., Kiguti L. R., Da Maia E. O., Fernandes L. T., Da Rocha L. R., Vilegas W., et al. (2013). Antidiarrheal and intestinal antiinflammatory activities of a methanolic extract of qualea parviflora mart. in experimental models. J. Ethnopharmacol. 150, 1016–1023. 10.1016/j.jep.2013.10.006 [DOI] [PubMed] [Google Scholar]
- Men X., Ma J., Wu T., Pu J., Wen S., Shen J., et al. (2018). Transcriptome profiling identified differentially expressed genes and pathways associated with tamoxifen resistance in human breast cancer. Oncotarget 9, 4074–4089. 10.18632/oncotarget.23694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. A., Hyland M., Ofner-Agostini M., Gourdeau M., Ishak M., Canadian Hospital Epidemiology Committee. Canadian Nosocomial Infection Surveillance, P. (2002). Morbidity, mortality, and healthcare burden of nosocomial clostridium difficile-associated diarrhea in Canadian hospitals. Infect. Control Hosp. Epidemiol. 23, 137–140. 10.1086/502023 [DOI] [PubMed] [Google Scholar]
- Nakamuta M., Kohjima M., Morizono S., Kotoh K., Yoshimoto T., Miyagi I., et al. (2005). Evaluation of fatty acid metabolism-related gene expression in nonalcoholic fatty liver disease. Int. J. Mol. Med. 16, 631–635. 10.3892/ijmm.20.3.351 [DOI] [PubMed] [Google Scholar]
- Pandey G., Gupta S. S., Bhatia A., Sidhu O. P., Rawat A. K., Rao C. V. (2017). Grilling enhances antidiarrheal activity of Terminalia bellerica Roxb. fruits. J. Ethnopharmacol. 202, 63–66. 10.1016/j.jep.2016.12.003 [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, 2002–2007. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarsini K. I., Khopde S. M., Kumar S. S., Mohan H. (2002). Free radical studies of ellagic acid, a natural phenolic antioxidant. J. Agric. Food Chem. 50, 2200. 10.1021/jf011275g [DOI] [PubMed] [Google Scholar]
- Puleston J., Cooper M., Murch S., Bid K., Makh S., Ashwood P., et al. (2005). A distinct subset of chemokines dominates the mucosal chemokine response in inflammatory bowel disease. Aliment. Pharm. Ther. 21, 109–120. 10.1111/j.1365-2036.2004.02262.x [DOI] [PubMed] [Google Scholar]
- Qu K., Wang Z., Tang K., Zhu Y., Fan R. (2019). Trehalose suppresses cadmium-activated Nrf2 signaling pathway to protect against spleen injury. Ecotoxicol. Environ. Saf. 181, 224–230. 10.1016/j.ecoenv.2019.06.007 [DOI] [PubMed] [Google Scholar]
- Ramirez N. M., Toledo R. C. L., Moreira M. E. C., Martino H. S. D., Benjamin L. D. A., De Queiroz J. H., et al. (2017). Anti-obesity effects of tea from Mangifera indica L. leaves of the Uba variety in high-fat diet-induced obese rats. Biomed. Pharmacother. 91, 938–945. 10.1016/j.biopha.2017.05.015 [DOI] [PubMed] [Google Scholar]
- Robert F. S., Rao J. P. (1996). Evidence for possible involvement of guanylate cyclase in diarrhoea induced by castor oil in mice. J. Diarrhoeal Dis. Res. 14, 218–219. 10.2307/23498666 [DOI] [PubMed] [Google Scholar]
- Sarnelli G., Seguella L., Pesce M., Lu J., Gigli S., Bruzzese E., et al. (2018). HIV-1 Tat-induced diarrhea is improved by the PPARalpha agonist, palmitoylethanolamide, by suppressing the activation of enteric glia. J. Neuroinflammation 15 (1), 94. 10.1186/s12974-018-1126-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q. X., Wang W., Chen M. H., Zhang H. F., Xu S. W. (2019). Ammonia induces Treg/Th1 imbalance with triggered NF-κB pathway leading to chicken respiratory inflammation response. Sci. Total Environ. 659, 354–362. 10.1016/j.scitotenv.2018.12.375 [DOI] [PubMed] [Google Scholar]
- Silverstein R. L., Febbraio M. (2009). CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal 2, re3. 10.1126/scisignal.272re3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H., Arora R., Arora S., Singh B. (2017). Ameliorative potential of Alstonia scholaris (Linn.) R. Br. against chronic constriction injury-induced neuropathic pain in rats. BMC Complement Altern Med. 17, 63. 10.1186/s12906-017-1577-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaria T., Bachli E., Schoedon G. (2019). Gene ontology analysis for drug targets of the whole genome transcriptome of human vascular endothelial cells in response to proinflammatory IL-1. Front. Pharmacol. 10, 414. 10.3389/fphar.2019.00414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stables M. J., Shah S., Camon E. B., Lovering R. C., Newson J., Bystrom J., et al. (2011). Transcriptomic analyses of murine resolution-phase macrophages. Blood 118, e192–e208. 10.1182/blood-2011-04-345330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Wei H., Bi Y., Wang Y., Zhao P., Zhang R., et al. (2018). Pre-cold acclimation improves the immune function of trachea and resistance to cold stress in broilers. J. Cell Physiol. 234, 7198–7212. 10.1002/jcp.27473 [DOI] [PubMed] [Google Scholar]
- Verotta L., Panzella L., Antenucci S., Calvenzani V., Tomay F., Petroni K., et al. (2018). Fermented pomegranate wastes as sustainable source of ellagic acid: antioxidant properties, anti-inflammatory action, and controlled release under simulated digestion conditions. Food Chem. 246, 129–136. 10.1016/j.foodchem.2017.10.131 [DOI] [PubMed] [Google Scholar]
- Vrese M. D., Offick B. (2010). Chapter 14–Probiotics and Prebiotics: Effects on Diarrhea. Bioactive Foods Promoting Health 137, 205–227. 10.1089/jmf.2005.067 [DOI] [Google Scholar]
- Wang S., Chi Q., Hu X., Cong Y., Li S. (2019). Hydrogen sulfide-induced oxidative stress leads to excessive mitochondrial fission to activate apoptosis in broiler myocardia. Ecotoxicol. Environ. Saf. 183, 1–9. 10.1016/j.ecoenv.2019.109578 [DOI] [PubMed] [Google Scholar]
- Wolff C., Boqvist S., Stahl K., Masembe C., Sternberg-Lewerin S. (2017). Biosecurity aspects of cattle production in Western Uganda, and associations with seroprevalence of brucellosis, salmonellosis and bovine viral diarrhoea. BMC Vet. Res. 13, 382. 10.1186/s12917-017-1306-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Chen S., Yu J., Li Y., Zhang X. Y., Yang L., et al. (2018). Single-cell transcriptome analyses reveal molecular signals to intrinsic and acquired paclitaxel resistance in esophageal squamous cancer cells. Cancer Lett. 420, 156–167. 10.1016/j.canlet.2018.01.059 [DOI] [PubMed] [Google Scholar]
- Yang Z. T., Zhou E. S., Wei D., Li D. P., Wei Z. K., Zhang W., et al. (2014). Emodin inhibits LPS-induced inflammatory response by activating PPAR-gamma in mouse mammary epithelial cells. Int. Immunopharmacol. 21, 354–360. 10.1016/j.intimp.2014.05.019 [DOI] [PubMed] [Google Scholar]
- Yao L., Wang J., Li B., Meng Y., Ma X., Si E., et al. (2018). Transcriptome sequencing and comparative analysis of differentially-expressed isoforms in the roots of Halogeton glomeratus under salt stress. Gene 646, 159–168. 10.1016/j.gene.2017.12.058 [DOI] [PubMed] [Google Scholar]
- Yaribeygi H., Mohammadi M. T., Sahebkar A. (2018). PPAR-alpha agonist improves hyperglycemia-induced oxidative stress in pancreatic cells by potentiating antioxidant defense system. Drug Res. (Stuttg) 68, 355–360. 10.1055/s-0043-121143 [DOI] [PubMed] [Google Scholar]
- Yim N. H., Gu M. J., Hwang Y. H., Cho W. K., Ma J. Y. (2016). Water extract of galla rhois with steaming process enhances apoptotic cell death in human colon cancer cells. Integr. Med. Res. 5, 284–292. 10.1016/j.imr.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuce A., Atessahin A., Ceribasi A. O., Aksakal M. (2007). Ellagic acid prevents cisplatin-induced oxidative stress in liver and heart tissue of rats. Basic Clin. Pharmacol. Toxicol 101, 345–349. 10.1111/j.1742-7843.2007.00129.x [DOI] [PubMed] [Google Scholar]
- Zeinali M., Rezaee S. A., Hosseinzadeh H. (2017). An overview on immunoregulatory and anti-inflammatory properties of chrysin and flavonoids substances. Biomed. Pharmacother. 92, 998–1009. 10.1016/j.biopha.2017.06.003 [DOI] [PubMed] [Google Scholar]
- Zhao S. S., Ma D. X., Zhu Y., Zhao J. H., Zhang Y., Chen J. Q., et al. (2018). Antidiarrhealeffect of bioactivity-guided fractions and bioactive components of pomegranate (Punica granatum L.) peels. Neurogastroenterol. Motil. : Off. J. Eur. Gastrointestinal Motil. Soc. 30, e13364–e13364. 10.1111/nmo.13364 [DOI] [PubMed] [Google Scholar]
- Zimmerman C. M., Bresee J. S., Parashar U. D., Riggs T. L., Holman R. C., Glass R. I. (2001). Cost of diarrhea-associated hospitalizations and outpatient visits in an insured population of young children in the united states. Pediatr. Infect. Dis. J. 20, 14–19. 10.1097/00006454-200101000-00004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated for this study can be found in NCBI GEO accession GSE142063.