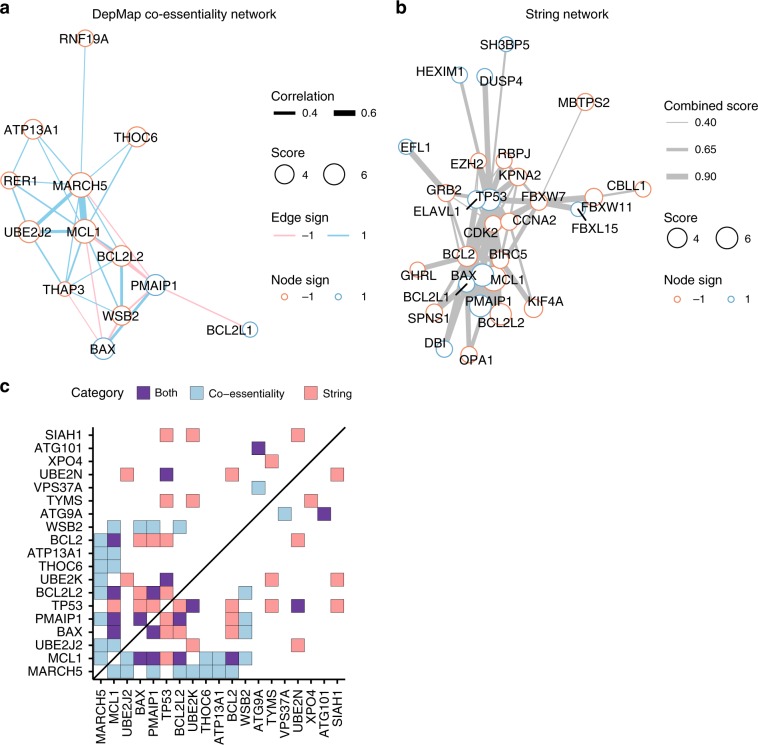
Fig. 3. BCL2L1 anchor screens reveal functionally coherent clusters of genes.
a Cluster of top hits from the DepMap co-essentiality network. Nodes represent genes and the size of each node is proportional to its average Z-score across all screens. Genes with an absolute average Z-score greater than 2 across BCL2L1 conditions are included in the network. Edges represent Pearson correlations across co-essentiality profiles in DepMap. Edges are drawn between genes with an absolute correlation greater than 0.2. Clustering was done by modularity optimization and a single cluster was chosen for visualization. b Cluster of top hits from the STRING network. Nodes are the same as a. Edges represent combined score in STRING. Edges are drawn between genes with a STRING combined score greater than 0.4. Clustering was done the same as a. c Interactions from DepMap and STRING between the top 20 hits by absolute Z-score. Genes are ordered by absolute average Z-score. Edge cutoffs are the same as a and b.