Abstract
Among more than 100 types of identified RNA modification, N6-methyladenosine (m6A) modification is the predominant mRNA modification, which regulates RNA splicing, translocation, stability, and translation. m6A modification plays critical roles in the growth, differentiation, and metabolism of cells. As a dynamic and reversible modification, m6A is catalyzed by “writers” (RNA methyltransferases), removed by “erasers” (demethylases), and interacts with “readers” (m6A-binding proteins). With more advanced technology applied to research, the molecular mechanisms of RNA methyltransferase, demethylase, and m6A-binding protein have been revealed. An increasing number of studies have implicated the correlation between m6A modification and human cancers. In this review, we summarize that the occurrence and development of various human cancers are associated with aberrant m6A modification. We also discuss the progress in research related to m6A modification, providing novel therapeutic insight and potential breakthrough in anticancer therapy.
Keywords: m6A, epigenetics, cancer, tumor suppressor
Background
N6-adenosine is the most prevalent epigenetic modification in RNA, originally identified in mRNAs in the 1970s.1 Similar to the methylation of DNA, N6-methyladenosine (m6A) methylation regulates post-transcriptional expression without changing the base sequence. m6A is the most abundant internal RNA modification, enriched at the RRACH motif in 3′ UTRs, 5′ UTRs, and near stop codons.2, 3, 4, 5, 6 It modulates RNA processing and metabolism, including alternative splicing, transport, translation, and degradation.7 As a dynamic and reversible modification, m6A deposition is regulated by methylases and demethylases. m6A-binding proteins subsequently recognize and bind the m6A-rich domain, inducing decay or accelerating translation efficiency. With the advancement of methods for detecting m6A and proteins, many enzymes have been identified and the functions and mechanisms of them have gradually emerged. Some studies looking into the mechanism of alternative splicing regulation have uncovered the function of m6A. m6A near splice sites in nascent pre-mRNA mediates heterogeneous nuclear ribonucleoprotein G (hnRNPG) binding. hnRNPG interacts with RNA polymerase II (RNAPII) with Arg-Gly-Gly (RGG) motifs, thereby modulating RNAPII occupancy and alternative splicing.8 Moreover, the location of m6A on nascent RNA is likely to modulate splicing kinetics. m6A co-transcriptionally depositing near splice junctions promotes fast splicing, while m6A in introns indicates long, slowly processed introns and alternative splicing events.9 m6A was also found in precursor mRNA (pre-mRNA), tRNA, microRNAs (miRNAs), and long non-coding RNAs (lncRNAs) later, but the functions of m6A in circular RNA (circRNA) remain elusive.10, 11, 12, 13, 14. m6A in circRNAs are frequently derived from exons that are not methylated in mRNAs, and a single m6A modification is enough to initiate circRNA translation, although the “writers” and “readers” are the same as those that participate in m6A modification in mRNA.15,16 With specific m6A-related mechanism, circRNA is involved in tumorigenesis, which indicates the important role of circRNA in human cancer.17
With burgeoning medical technology, substantial progress has been made in the detection and diagnosis of cancer. Accumulating studies indicate that aberrant m6A levels closely correlate with carcinogenesis and the progression and metastasis of cancer cells. The dysregulation of writers, “erasers,” and readers is proved to be the culprit, activating oncogenes or inhibiting tumor suppressor genes by activating signaling pathways.18,19 Potential therapeutic targets have also been offered by researching the mechanisms of carcinogenesis. In this review, we discuss the relationships between human cancers and new discoveries in the regulation of m6A modification.
Regulation of m6A Modification
Methyltransferases
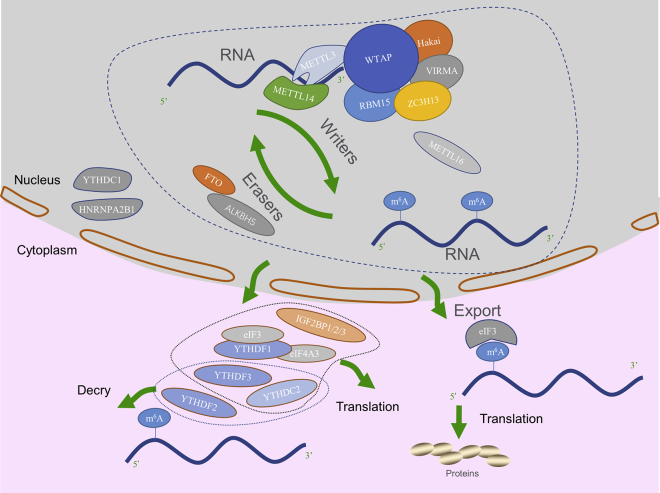
m6A writers (methyltransferases) can install the m6A RNA modification (Figure 1). METTL3 is the first known RNA m6A methyltransferase. Then, METTL14 was identified, forming a stable METTL3-METTL14 complex that is also called the m6A-METTL complex (MAC).20 The interface of the heterodimer is formed by strands β4–5 and helix α4 of each methyltransferase domain (MTD), and it contains hydrogen bonds as well as hydrophobic interactions. The hydrophobic region centering on strands β4–5 is protected from solvent exposure by an N-terminal extension of METTL14. In addition, METTL3 and METTL14 each forms a partially disordered loop that can insert an aromatic residue into a hydrophobic pocket in the other subunit. All of these unique structural features enhance the stability and compactness of MAC.14,21 METTL3 is the only catalytic subunit of MAC, while METTL14 maintains MAC integrity and is likely to mediate RNA binding. Sequence analysis indicated that the METTL3 catalytic site contains a more conserved DPPW motif, whereas the catalytic motif of METTL14 is a divergent EPPL sequence. Moreover, two Cys-Cys-Cys-His (CCCH)-type zinc fingers of METTL3, adjacent to MTD3 and connected by an anti-parallel β sheet, are also necessary for methylation activity. The zinc finger domain (ZFD) specifically recognizes the 5′-GGACU-3′ consensus sequence of RNA, forming an RNA-binding interface. Although METTL14 has the folding configuration similar to METTL13, the cavity where RNA substrates bind is not possessed.22,23 However, recent research has uncovered the important role of METTL14 in the crosstalk between histone modification and RNA methylation. METTL14 recognizes and directly binds with histone H3 trimethylation at Lys36 (H3K36me3), facilitating MAC to adjacent RNAPII, thereby installing m6A in actively transcribed nascent RNAs co-transcriptionally. This mechanism may suggest how m6A is specifically deposited in the transcriptome.24
Figure 1.
Methyltransferases, Demethylases, and m6A-Binding Proteins in m6A Modification
m6A is catalyzed by writers and removed by erasers (FTO and ALKBH5). MACOM, consisting of WTAP, VIRMA, Hakai, RBM15, and ZC3H13, ensures the location of the METTL3-METTL14 core complex to nuclear speckle. m6A modification exerts biological functions by binding to YTHDC1–2, YTHDF1–3, HNRNPA2B1, IGF2BP1–3, eIF3, and eIF4A3. In the nucleus, YTHDC1 and HNRNPA2B1 bind m6A and perform multiple functions. In the cytoplasm, YTHDC2, YTHDF1–3, IGF2BP1–3, eIF3, and eIF4A3 induce translation or degradation of transcripts. eIF3 can promote translation by directly binding m6A in the 5′ UTR.
The discovery of Wilms’ tumor 1-associating protein (WTAP) enriched the composition of the m6A methyltransferase complex. WTAP exhibits affinity for the methyltransferase complex, locating the METTL3-METTL14 complex at nuclear speckles and recruiting them to mRNA targets, regulating expression and alternative splicing of genes with the assistance of METTL3.25 Studies focusing on interaction surfaces within the METTL3-METTL14-WTAP complex unveiled the novel overall architecture. In WTAP, the METTL3-binding surface is within the N-terminal 150 aa, while the WTAP interaction surface on METTL3 is an N-terminal helical structure that is necessary and sufficient for WTAP-METTL3 interaction. Strikingly, METTL3 phosphorylation in the WTAP interaction surface does not affect subcellular localization, WTAP interaction, or catalytic activity. Moreover, the C-terminal arginine-glycine repeats (RGG) of METTL14, contributing to RNA substrate binding, are indispensable for MAC catalytic activity. The finding is consistent with previous conclusions about the function of METTL14.26
More WTAP-related proteins and their relevant regulatory factors have been discovered in further studies. Hitherto, we defined the complex consisting of WTAP, VIRMA, Hakai, RBM15 (RNA-binding motif protein 15), and ZC3H13 (a zinc-finger protein) as MACOM (m6A-METTL-associated complex).11,27, 28, 29 Interacting with each other, these factors accumulate around target RNAs and catalyze methylation in specific sites. WTAP plays a central role in MACOM and mediates the nuclear speckle localization of the complex with assistance of BCLAF1 and THRAP3 (arginine/serine-rich domain-containing proteins).30
Recently, METTL16 was regarded as another human m6A methyltransferase that targets pre-mRNAs and non-coding RNAs. METTL16 participates in splicing regulation by catalyzing N6-methylation in A43, which is in the specific sequence of U6 small nuclear RNA (snRNA). The alteration in A43 can influence the base pairing at 5′ splice sites of pre-mRNAs during splicing.13 Intriguingly, studies also revealed METTL16 as a regulator that maintains SAM homeostasis by binding with MAT2A mRNA hairpins, implicating its likely important role in early development.31,32
Demethylases
Termed as erasers, demethylases can remove m6A in RNA. To date, only two demethylases, fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5), have been identified. FTO is the first identified demethylase, which is highly expressed in brain and muscle.33 FTO mediates demethylation with its oxidative activity targeting the m6A-rich region in RNA.34 The mechanism of sequence-specific m6A demethylation was revealed by further studies that demonstrated that FTO and RCas9 can fuse together as an RNA-targeting module. The resulting RCas9-FTO retained demethylation activity and bound to RNA in a sequence-specific manner depending on the single-guide RNA (sgRNA) and PAMmer.35 Furthermore, consensus motifs GGACU and RRACU were discovered in target sequences. FTO specifically removed m6A from GGACU and RRACU motifs in a concentration-dependent manner.4 Additionally, FTO also mediates the demethylation of cap N6, 2′-O-dimethyladenosine (cap m6Am) in snRNA and mRNA. Compared with the demethylation of m6A, FTO act differently in m6Am demethylation, which can be accounted for by different cellular distribution features of FTO. Experiments conducted in polyadenylated RNAs among different cell lines manifested that FTO is more active with regard to m6Am (81.9% of m6Am and 20.3% of m6A were demethylated in vitro). m6A is preferentially demethylated by FTO in nucleus, whereas cap m6Am is a prominent target in cytoplasm.36 The cap m6Am at +1 position from 5′ cap in mRNA confers resistance to mRNA-decapping enzyme DCP2 on transcripts, thereby enhancing mRNA stability.37 FTO-mediated m6Am demethylation was also detected in snRNAs (U1 RNA and U2 RNA; both the cap and internal m6Am modifications are more significant in U2 RNA), but m6A demethylation only happened in U6 RNA.36 Demethylation of m6Am at the sites adjacent to snRNA cap lead to relatively decreased m6Am-snRNA levels. However, the function of FTO can be inhibited by the oncometabolite-2-hydroxyglutarate, resulting in increased m6Am-snRNA levels, which may change patterns of alternative splicing.38 The FTO-induced noteworthy changes of snRNAs indicate that FTO is capable of having an influence on mRNA splicing.
ALKBH5 catalyzes demethylation of m6A in RNA and contributes to normal splicing and the formation of longer 3′ UTR mRNAs. Therefore, ALKBH5 also takes part in maintaining stability of transcripts.33,39 In light of the similar functions of ALKBH5 and FTO, the underlying mechanism by which the demethylases selectively recognize their target transcripts is intriguing. Some research has indicated that an m6A-induced conformational change on RNA may account for the specificity but that the consensus sequence (GG(m6A)CU) is not indispensable for the specific recognition.40
m6A-Binding Proteins
m6A modification exerts biological functions by binding to YTH domain-containing proteins (YTHDC1–2), YTH-family proteins (YTHDF1–3), and other interacting factors. The proteins are collectively defined as readers. In the nucleus, YTHDC1 promotes exon inclusion of targeted mRNAs and regulates mRNA splicing and export from nucleus to cytoplasm by recruiting pre-mRNA splicing factor SFSF3.41,42 YTHDC1 was also established to participate in the maintenance of intracellular SAM levels, thereby regulating the level of methylation.32 HNRNPA2B1 is a direct m6A reader which takes part in primary RNA (pri-miRNA) processing and alternative splicing by interacting with the DGCR8 protein.43 In the cytoplasm, YTHDF1–3 proteins work synergistically to influence RNA metabolism.41,44,45 YTHDF3 promotes translation initiation of its target mRNA with assistance of YTHDF1 and initiation factor eIF4A3. YTHDF3 also mediates mRNA degradation by cooperating with YTHDF2. Similar to YTHDF3, YTHDC2 also has a dual effect on target mRNA. YTHDC2 enhances translation efficiency of target mRNA, while knockdown of YTHDC2 results in an upregulation of m6A-modified transcripts. This novel function of YTHDC2 is essential for fertility in mammals and ensures the transition from mitosis to meiosis.46,47 Especially, transcripts with m6A in the 5′ UTR can be translated by directly binding elF3, independent of YTHDF1 and cap-binding factor eIF4E. In addition, diverse cellular stresses selectively result in an increase in mRNAs with 5′ UTR m6A.5
As a new class of m6A readers discovered recently, IGF2BP1/2/3 (insulin-like growth factor 2 mRNA-binding proteins 1, 2, and 3) recognize the consensus GG(m6A)C sequence and bind m6A by their K homology domains. In such an m6A-dependent manner, IGF2BP1/2/3 prevent target mRNAs from degradation and promote mRNA translation.48
m6A Modification in the Occurrence and Development of Cancers
Acute Myeloid Leukemia (AML)
AML is a malignant disease originating from hematopoietic stem cells or progenitor cells. Chemotherapy and hematopoietic stem cell transplantation are general treatments for AML, but targeted therapy, demethylation therapy, and immunotherapy also show promising therapeutic effects. With high expression of methyltransferases, AML cells feature elevated m6A, which contributes to the maintenance of multilineage differentiation potential and inhibits cell differentiation in AML.49,50 METTL3 recruited by CEBPZ promotes translation of SP1 by upregulating the m6A level. SP1 subsequently activates the oncogene c-MYC, which can result in the development of AML.50 Similarly, m6A-modified BCL2 and PTEN induced by elevated METTL3 levels in AML can also lead to AML development caused by downregulation of phosphorylated phosphatidylinositol 3-kinase (PI3K)/AKT.49 However, elevated non-functional METTL3 also activates the PI3K/AKT pathway, which implicated that there are certain mechanisms independent of m6A modification.49 Strikingly, similar to m6A readers, METTL3 can also promote the translation of mRNAs by binding m6A-modified regions close to the stop codon and interacting with eIF3h.51 METTL14 is essential for maintaining self-renewal of AML cells, while knockdown of METTL14 promotes myeloid differentiation. The consistent high expression of METTL14 is attributed to the dysregulation of SPI1.52 Oncogenes MYB and MYC are targets of METTL14. However, YTHDF proteins are not involved in promoting stability and translation of target mRNAs, while knockdown of METTL14 has an effect on the quantity of MYB and MYC mRNA.52 METTL16 indirectly regulates the activity of METTL3/MELL14 by regulating the expression of MAT2A, which maintains appropriate SAM levels.32,53
WTAP has been found upregulated in AML and to plays an oncogenic role.54 Increased expression of WTAP in AML cell lines alters alternative splicing, promotes proliferation, and blocks differentiation.30,54 Studies found that the increased WTAP protein levels result from simultaneous high levels of cytoplasmic METTL3, in agreement with the conclusion that METTL3 and METTL14 are WTAP complex interactors.55
The RBM15-MKL1 fusion was originally detected in infants and the transcripts were analyzed for diagnosis of AMKL.56,57 Interacting with the KMT2G, RBM15-MKL1 fusion protein enhanced cell proliferation.58 RBM15 can inhibit myeloid differentiation in hematopoietic cells by stimulating Notch signaling via RBPJkappa59 and contribute to adult hematopoiesis and normal megakaryocyte development mediated at least in part by c-Myc. Therefore, the function of RBM15 in AML is correlated with the activation of Notch signaling and interaction with c-Myc.60 In addition, overexpression of PRMT1 leads to aberrant alternative splicing and blocks AML cell differentiation by inducing RBM15 ubiquitylation and degradation.61,62
FTO is overexpressed in AML with MLL rearrangements, PML-RARA, FLT3-ITD, or NPM1 mutations.63 FTO can decrease m6A levels in certain mRNA transcripts, including the tumor suppressor ankyrin repeat and ASB2 (SOCS box protein 2) and RARA (retinoic acid receptor alpha), thereby suppressing the expression of ASB2 and RARA, which regulate normal hematopoiesis and all-trans-retinoic acid (ATRA)-induced AML cell differentiation. Therefore, overexpressed FTO mediates leukemogenesis and indicates better efficacy of ATRA treatment in AML cells.63 With the capacity for catalyzing demethylation of m6Am in mRNA and snRNA and N1-methyladenosine (m1A) in tRNA,36 FTO may mediate oncogenesis independent of m6A. Similarly, CAPAM (cap-specific adenosine methyltransferase) mediating m6A modification at the 5′ cap in mRNA may correlate with AML, but the hypothesizes have yet to be identified.64,65
Recently, an investigation in leukemia patients indicated that genetic alterations of m6A regulatory genes were associated with p53 mutations in AML. These alterations always predict a poor survival rate.66 Genetic aberrations, that is, CBFA2T3/GLIS2 and NUP98/KDM5A, may be used for risk group stratification of pediatric AMKL and treatment tailoring.67
Colorectal Cancer
YTHDF1 and METTL3 have been identified to play significant roles in colorectal cancer (CRC). YTHDF1 was established to regulate tumorigenesis in CRC cells via the Wnt/β-catenin pathway. Mechanistically, studies found that YTHDF1 induced aberrant activation of Wnt/β-catenin signaling via recognizing and promoting translation of FZD9 and Wnt6 mRNA in an m6A-dependent manner. According to clinical and laboratory data, high YTHDF1 expression in patients signaled poorer overall survival, while silencing of YTHDF1 in vitro resulted in poor efficacy of anticancer drugs and suppression of cancer proliferation. However, it is notable that c-Myc was indispensable in the carcinogenic mechanism of YTHDF1, indicating the theory of a c-Myc-driven YTHDF1 axis.68 Moreover, YTHDF1 gene copy number amplification contributes to YTHDF1 overexpression in CRC.69 METTL3, acting as a carcinogenic factor, was observed to be overexpressed in CRC metastatic tissues and released a signal of poor prognosis. Confirmed as an m6A methyltransferase (as previously mentioned), METTL3 is capable of methylating SRY (sex determining region Y)-box 2 (SOX2), which is essential for maintaining self-renewal. IGF2BP2 subsequently recognizes the methylated SOX2 transcripts, thereby preventing SOX2 mRNA degradation.70
In addition, studies have shown that lncRNA RP11 triggers the dissemination of CRC cells. RP11 induce cell dissemination by post-translationally regulates the stability of Zeb1, an epithelial-mesenchymal transition (EMT)-related transcription factor. An m6A-dependent manner is involved in the accumulation of RP11 in nuclear. Mechanistically, the RP11/HNRNPA2B1/mRNA complex prevented the proteasomal degradation of Zeb1 by promoting the mRNA degradation of Siah1 and Fbxo45.71
Gastric Cancer
The molecular mechanism in the occurrence of gastric cancer (GC) remains unclear, but some conclusions have emerged during studies. Some studies have given way to the idea that METTL3 is a carcinogenic factor of GC. METTL3 was observed to be significantly overexpressed in GC tissues, and the expression level of METTL3 was positively related to the tumor/node/metastasis (TNM) stage of GC. m6A methylation level was also upregulated simultaneously. METTL3 leads to inactivation of the AKT signaling pathway, while knockdown of METTL3 activated the apoptotic pathway in GC cells. Downregulation of METTL3 suppressed the proliferation and mobility of GC cells.72,73 However, another study came to the opposite conclusion that downregulation of m6A contributed to GC cell proliferation and invasiveness by activating Wnt and PI3K/AKT signaling, indicating that the m6A methyltransferases may suppress tumorigenesis.74 Moreover, compared with adjacent non-tumor tissues, FTO expression in GC tissues featured a high level in protein and mRNA, suggesting that FTO may play a significant role in GC. Overexpression of FTO promotes the proliferation, migration, and invasion of GC cells and is positively related to TNM stage.75
Breast Cancer
Aberrant m6A level and dysregulation of METTL3, METTL14, WTAP, and FTO have been observed in breast cancer (BC) cells.76 Decreased m6A level was confirmed to mediate abnormal expression of certain tumor suppressor genes in a premature polyadenylation (pPA)-dependent manner.77 Studies have shown that FTO was upregulated in human BC, promoting proliferation, colony formation, and metastasis of BC cells. Mechanistically, FTO inhibits the function of BNIP3, which acts as a tumor suppressor. FTO mediated m6A demethylation at the RRACH motif near the stop codon of BNIP3 mRNA and induced its degradation in an YTHDF2-independent manner.78 As a reader of the m6A, HNRNPA2/B1 is upregulated in BC cells, reducing sensitivity to antagonists of estrogen receptor by regulating levels of specific miRNAs.79 Specially, some studies found the dual role of m6A in BC. In immortalized and oncogenically transformed human mammary epithelial cells (HMECs), m6A levels were significantly decreased with a decrease of METTL3 and an increase of ALKBH5. However, increased m6A levels in transformed cells, resulting from overexpression of METTL3 and METTL14, or deletion of ALKBH5, promoted proliferation and migration. Interestingly, m6A levels in the immortalized and transformed cells were found to increase under stress with constant levels of METTL3, METTL14, and ALKBH5, suggesting other underlying pathways for regulation of m6A levels.80
Lung Cancer
Deregulation of FTO and METTL3 has been detected in lung cancer, which indicated their significant roles in tumorigenesis and development. Studies established that FTO plays an oncogenic role in NSCLC (non-small-cell lung cancer) and LUSC (lung squamous cell carcinoma). In human NSCLC tissues, overexpressed FTO promoted proliferation and colony formation of cancer cells by increasing USP7 mRNA level and USP7 expression.81 FTO was also involved in the progression of LUSC. Mechanistically, FTO decreased m6A levels and mRNA stability in the MZF1 mRNA transcript, thereby promoting expression of MZF1 and mediating carcinogenesis.82 METTL3-eIF3h enhances translation of oncogenic mRNAs in primary lung tumors and leads to oncogenic transformation.51 METTL3 is also implicated in NSCLC progression, promoting cell growth and survival via the PI3K/Akt pathway.83 Additionally, METTL16 was found to bind the 3′ terminal triple helix of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), a cancer-promoting long noncoding RNA, and regulated MALAT1 endocellular accumulation. However, the relationship between tumorigenesis and this mechanism remains unclear.84 In addition, when researching into the deregulated autophagy in human cancers, aberrant activation of the oncogene UBE2C was noticeable. In NSCLC, upregulated ALKBH5 maintains a lower m6A level in UBE2C mature RNAs and activates UBE2C expression, thereby repressing autophagy. Depletion of UBE2C induces attenuated proliferation, clonogenicity, and invasive growth of NSCLC cells.85
Hepatocellular Carcinoma
Overexpressed METTL3, METTL14 downregulation, and upregulated m6A readers have been observed in hepatocellular carcinoma (HCC) cells. Overexpression of METTL3 is a signal of poor prognosis in HCC, and upregulated YTHDF1 is positively correlated with TNM stage, indicating their important roles in HCC cells metastasis and progression.86, 87, 88 With a YTHDF2-dependent mechanism, METTL3 mediated m6A modification of the SOCS2 suppressor, thereby repressing SOCS2 expression and promoting HCC progression.86 Contrastingly, METTL14 performs a repressing effect in tumor metastasis by interacting with the microprocessor protein DGCR8 and promoting pre-miRNA126 process via m6A modification.89 VIRMA, another m6A writer, was also identified as an oncogenic factor, facilitating migration, proliferation, and invasion of HCC cells by inducing m6A modification of ID2 mRNA.90
Melanoma
Melanoma is a type of malignant cancer that originates from pigment-producing melanocytes. The pathogenesis of melanoma still remains unclear, but some studies have given insight into the mechanism of oncogenesis. FTO and METTL3 are significantly upregulated in human melanoma.91,92 FTO-mediated m6A mRNA demethylation promotes melanoma tumorigenesis and anti-PD-1 resistance. Knockdown of FTO increases m6A modification in the critical protumorigenic melanoma cell-intrinsic genes, including PD-1, CXCR4, and SOX10, inducing RNA degradation via binding YTHDF2. In mice, knockdown of FTO markedly enhances melanoma cell sensitivity to interferon (IFN)-γ and anti-PD-1 treatment, which suggested a promising anti-cancer therapy.91 Overexpressed METTL3 enhances colony formation and invasion of melanoma cells via promoting accumulation of MMP2 and N-cadherin.92
Glioblastoma (GBM)
Glioblastoma is the most aggressive malignant tumor of central nervous system. Although there are multiple clinical trials for GBM, the recurrence rate is still very high. GB stem cells (GSC) are a subset of GBM cells, closely correlating with tumorigenesis, proliferation, and recurrence. Many studies aiming at GSCs have been performed in recent years. Upregulated m6A mRNA modification was identified to induce the suppression of GSC self-renewal and tumorigenesis. Mechanistically, the changing m6A level altered mRNA expression of certain genes that play critical roles in GSCs. METTL3 and METTL14 suppress GSC growth and self-renewal by downregulating oncogenes (ADAM19, EPHA3, and KLF4) and upregulating tumor suppressors (CDKN2A, BRCA2, and TP53I11).93 Further studies have shed light on the function of some enzymes. ALKBH5 was overexpressed in GSCs and promoted FOXM1 expression by demethylating its nascent transcripts. Moreover, a lncRNA antisense to FOXM1 (FOXM1-AS) also participated in the ALKBH5-mediated tumorigenesis.94 Being involved in RNA processing and oncogenic pathways of GSCs, functions of METTL3 are multiple and complex. METTL3 plays a dominant role in m6A modification of GSCs and participates in the expression of GSC-specific actively transcribed genes and alternative splicing. Additionally, METTL3 decreases A-to-I RNA editing by downregulating ADAR and ADARB1, whereas it upregulates editing enzymes APOBEC1 and APOBEC3A to increase C-to-U RNA editing.95
Advances in the m6A Modification on Human Cancers
It is becoming increasingly clear that deregulated m6A modification is a key factor in growth, metastasis, and drug resistance of various tumors. Recently, some novel regulatory pathways have been found in cancer cells, which may give insight into potential therapeutic strategies to inhibit cancers. Many enzymes and genes have been identified to play vital roles in the sophisticated system and alter therapeutic efficacy.
METTL3 has been considered to be an oncogenic factor in many human cancers, and studies focusing on blocking METTL3-dependent pathways have been performed. METTL3-depleted pancreatic cancer cells exhibited higher sensitivity to anticancer reagents.96 miR-600-induced METTL3 suppression can inhibit NSCLC progression.83 In bladder cancer cells, METTL3 and YTHDF1 promote oncogene CDCP1 translation by binding m6A in the 3′ UTR of CDCP1 mRNA. The METTL3-m6A-CDCP1 axis has been revealed as a target for treating chemical-induced cancers.97 Moreover, METTL3 participates in the mutagenesis of p53 proteins, which promotes proliferation, metastasis, and drug resistance of cancer cells. METTL3 catalyzes the formation of m6A at the point-mutated codon 273 (G>A) of p53 pre-mRNA, promoting aberrant splicing of p53 pre-mRNA and expression of p53 R273H mutant protein. Furthermore, upregulated METTL3 expression is partly attributed to glycosphingolipids, which execute function via activating cSrc and β-catenin signaling.98 Reduction of m6A methylation caused by abnormal MAC has been found in about 70% of human endometrial cancer samples, resulting in activation of the AKT pathway by upregulating the negative AKT regulator PHLPP2 and downregulating the positive AKT regulator mTORC2. The alteration of MAC can be caused by METTL14 mutation (hotspot R298P mutation) or reduced expression of METTL3.99
FTO has been observed significantly upregulated in human cancers, promoting occurrence, proliferation, and migration of cancer cells. These discoveries have conferred an identity as a therapeutic target on FTO. FB23 and FB23-2 are promising small-molecule FTO inhibitors. FB23-2 negatively regulates proliferation and progression of human AML cell lines by executing inhibition on FTO.100 In the emerging FTO-m6A-MYC-CEBPA axis, FTO is the target of R-2HG (R-enantiomer of 2-hydroxyglutarate), which has anticancer activity in AML. R-2HG disabled FTO and upregulated m6A level in MYC/CEBPA mRNA, reducing the stability and translation of MYC and CEBPA transcripts. High expression of FTO improved sensitivity to R-2HG, whereas overload of m6A in MYC mRNA weakens the anticancer effect of R-2HG. Similarly, in R-2HG-sensitive leukemia cells, mutant IDH1R132H also induced cell-cycle arrest, apoptosis, and proliferation inhibition.101 Besides improving sensitivity to cancer suppressors, FTO also enhances resistance to chemoradiotherapy. In cervical squamous cell carcinoma (CSCC) tissues, FTO decreased the m6A level in β-catenin mRNA transcripts and regulated expression of β-catenin, thereby promoting activity of excision repair cross-complementation group 1 (ERCC1).102 However, FTO is suppressed in clear cell renal cell carcinoma (ccRCC) tissue, and the level of FTO is negatively correlated with tumor severity. This phenomenon is attributed to the FTO-PGC-1alpha axis. With demethylase activity, FTO removes m6A residues in PGC-1alpha mRNA transcripts and increases expression of PGC-1alpha, inducing oxidative stress and impaired tumor growth.103 In addition, clinical studies have demonstrated the relationship between tumor severity and the expression of FTO and ALKBH1. According to clinical statistics of GC patients, the expression level of ALKBH1 is negatively correlated with tumor sizes and TNM stages, while expression of FTO is associated with better overall survival.104
EMT, a crucial step for tumor metastasis, has been confirmed to involve an m6A/YTHDF1-mediated mechanism. YTHDF1 recognizes upregulated m6A in coding sequences of Snail (a key transcription factor of EMT), promoting translation of Snail mRNA.105 Studies focusing on durable neoantigen-specific immunity also uncovered the critical role of YTHDF1. In classical dendritic cells of mice, YTHDF1 interferes with cross-presentation of tumor antigens and cross-priming of CD8+ T cells. YTHDF1 binds the m6A-modified transcripts, which encode lysosomal proteases, promoting the translation of lysosomal cathepsins. Conversely, knockdown of YTHDF1 induces suppression of cathepsins and dramatically enhances cross-presentation. Furthermore, experiments in vivo implicated that YTHDF1 is a potential therapeutic target for enhancing efficacy of PD-L1 checkpoint blockade.106
What may bring breakthroughs for targeted therapy is the discovery of the cell specificity of YTHDF2 function. YTHDF2 is not essential for normal hematopoietic stem cell (HSC) function, whereas in leukemic stem cells, it blocks the apoptosis pathway by regulating expression of tumor necrosis factor receptor superfamily 2 (TNFRSF2).107 Suppression of YTHDF2 is promising for maintenance of HSC self-renewal capacity after transplantation. Knockdown of human YTHDF2 led to a significant increase of HSCs (an average increase of 14.3-fold and 13.6-fold in the ex vivo expansion of human umbilical cord blood [hUCB] HSCs, a 5.1-fold increase in colony-forming units [CFU], and more than an 8-fold increase in functional hUCB HSCs in the secondary serial of a limiting dilution transplantation assay), and hematopoietic malignancies were not detected in YTHDF2-knockout mice. Mechanically, YTHDF2 promotes decay of m6A-modified mRNAs, which are critical for stem cell self-renewal.108 Thus, YTHDF2 is identified as a unique therapeutic target.
Conclusion and Perspective
To summarize, overexpression or suppression of writers and erasers lead to aberrant levels of m6A in the transcripts of oncogenes or tumor suppressor genes, regulating translation efficiency and degradation with full participation of readers (Table 1). These resulting proteins induce activation of signaling pathways (Wnt/β-catenin, PI3K/AKT) and then create cascade reactions. However, we know little about how the enzymes specifically recognize the genes and transcripts.
Table 1.
m6A Methylation-Related Human Cancers
| Cancer | Molecule | Function | Mechanism | References |
|---|---|---|---|---|
| AML | METTL3 | oncogenic | (1) indirectly activates the oncogene c-MYC by upregulating m6A modification of SP1 and stimulates its translation (2) induces m6A modification of BCL2 and PTEN and then downregulates apoptosis and the PI3K/AKT pathway (3) promotes the translation of mRNAs by binding m6A-modified regions and interacting with eIF3h |
50 |
| 49 | ||||
| 51 | ||||
| METTL14 | oncogenic | activates the SPI1-METTL14-MYB/MYC signaling pathway | 52 | |
| METTL16 | oncogenic | regulates the activity of METTL3/MELL14 indirectly | 32,53 | |
| FTO | oncogenic | (1) decreases m6A levels in mRNA transcripts of the tumor suppressors ASB2 and RARA, thereby suppressing the expression of them (2) mediates oncogenesis independent of m6A |
63 | |
| 36 | ||||
| WTAP | oncogenic | promotes proliferation and blocks differentiation of AML cells | 30,54 | |
| RBM15 | oncogenic | (1) enhances cell proliferation via RBM15-MKL1 fusion (2) inhibits myeloid differentiation by stimulating Notch signaling via RBPJkappa (3) blocks AML cell differentiation via the PRMT1-RBM15 axis |
56, 57, 58 | |
| 59 | ||||
| 61,62 | ||||
| CRC | YTHDF1 | oncogenic | induces aberrant activation of Wnt/β-catenin signaling by recognizing and promoting the translation of m6A-modified FZD9 and Wnt6 mRNA | 68 |
| METTL3 | oncogenic | maintains SOX2 expression through an m6A-IGF2BP2-dependent mechanism | 70 | |
| RP11 | oncogenic | induces cell dissemination via post-translational upregulation of Zeb1 | 71 | |
| GC | METTL3 | oncogenic suppressive |
promotes proliferation and mobility of GC cells by inactivating the AKT signaling pathway | 72,73 |
| suppresses proliferation and invasiveness by inactivating Wnt and PI3K‐Akt signaling | 74 | |||
| FTO | oncogenic | unclear | 75 | |
| BC | FTO | oncogenic | inhibits tumor suppressor BNIP3 | 78 |
| HNRNPA2/B1 | oncogenic | reduces BC cell sensitivity to antagonists of estrogen receptor | 79 | |
| Lung cancer | METTL3 | oncogenic | (1) promotes growth and survival of NSCLC cells via the PI3K/Akt pathway (2) promotes translation of oncogenic mRNAs in a METTL3-eIF3h manner in primary lung tumors |
83 |
| 51 | ||||
| METTL16 | unclear | binds the 3′ terminal triple helix of MALAT1 | 84 | |
| FTO | oncogenic | (1) promotes proliferation and colony formation of NSCLC cells by increasing the expression of USP7 (2) mediates carcinogenesis in LUSC by promoting expression of MZF1 |
81 | |
| 82 | ||||
| ALKBH5 | oncogenic | activates oncogene UBE2C expression | 85 | |
| HCC | METTL3 | oncogenic | represses SOCS2 expression in HCC through an m6A-YTHDF2-dependent mechanism | 86, 87, 88 |
| METTL14 | suppressive | interacts with DGCR8 and promotes primary miRNA126 processing in an m6A-dependent manner | 89 | |
| VIRMA | oncogenic | induces m6A modification of ID2 mRNA | 90 | |
| Melanoma | METTL3 | oncogenic | promotes accumulation of MMP2 and N-cadherin and enhances colony formation and invasion | 92 |
| FTO | oncogenic | increases RNA degradation of PD-1, CXCR4, and SOX10 via binding YTHDF2 | 91 | |
| GBM | METTL3/METTL14 | suppressor | suppresses GSC growth and self-renewal by downregulating oncogenes (ADAM19, EPHA3, and KLF4) and upregulating tumor suppressors (CDKN2A, BRCA2, and TP53I11) | 93 |
| ALKBH5 | oncogenic | promotes FOXM1 expression | 94 | |
| Endometrial cancer | METTL3/METTL14 | suppressor | reduces m6A methylation, upregulates the negative AKT regulator PHLPP2, and downregulates the positive AKT regulator mTORC2, thereby activating the AKT pathway | 99 |
By detecting the expression levels of the enzymes in cancerous tissues and adjacent non-cancerous tissues, studies have shown the prevalent overexpression of METTL3, METTL14, FTO, and others. However, METTL3 was identified as a suppressor in certain cancers, which is in conflict with previous conclusions. Further studies have indicated that the m6A level was changing with the development of cancer, which may partly explain the contradiction. The exact regulatory mechanism of m6A-related protein expression is not fully clear. m6A modification provides a new idea for cancer therapy. Further studies are needed for weakening drug resistance of cancer cells and improving the accuracy of targeted therapy.
Author Contributions
L.L., Y.W., and J.W. wrote and drafted the manuscript and figures. H.F., Z.Q., and J.L., revised the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant/award nos. 81701019 and 81870763).
Contributor Information
Jingwen Liu, Email: liujingwen@tmu.edu.cn.
Zongchang Qin, Email: wyw@qdu.edu.cn.
Hong Fan, Email: afanhong@126.com.
References
- 1.Desrosiers R., Friderici K., Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roundtree I.A., Evans M.E., Pan T., He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y., Wu K., Quan W., Yu L., Chen S., Cheng C., Wu Q., Zhao S., Zhang Y., Zhou L. The dynamics of FTO binding and demethylation from the m6A motifs. RNA Biol. 2019;16:1179–1189. doi: 10.1080/15476286.2019.1621120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.B., Jaffrey S.R. 5′ UTR m6A promotes cap-independent translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun T., Wu R., Ming L. The role of m6A RNA methylation in cancer. Biomed. Pharmacother. 2019;112:108613. doi: 10.1016/j.biopha.2019.108613. [DOI] [PubMed] [Google Scholar]
- 7.Lan Q., Liu P.Y., Haase J., Bell J.L., Hüttelmaier S., Liu T. The critical role of RNA m6A methylation in cancer. Cancer Res. 2019;79:1285–1292. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 8.Zhou K.I., Shi H., Lyu R., Wylder A.C., Matuszek Ż., Pan J.N., He C., Parisien M., Pan T. Regulation of co-transcriptional pre-mRNA splicing by m6A through the low-complexity protein hnRNPG. Mol. Cell. 2019;76:70–81.e9. doi: 10.1016/j.molcel.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louloupi A., Ntini E., Conrad T., Ørom U.A.V. Transient N-6-methyladenosine transcriptome sequencing reveals a regulatory role of m6A in splicing efficiency. Cell Rep. 2018;23:3429–3437. doi: 10.1016/j.celrep.2018.05.077. [DOI] [PubMed] [Google Scholar]
- 10.Alarcón C.R., Lee H., Goodarzi H., Halberg N., Tavazoie S.F. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patil D.P., Chen C.K., Pickering B.F., Chow A., Jackson C., Guttman M., Jaffrey S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X.Y., Zhang J., Zhu J.S. The role of m6A RNA methylation in human cancer. Mol. Cancer. 2019;18:103. doi: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warda A.S., Kretschmer J., Hackert P., Lenz C., Urlaub H., Höbartner C., Sloan K.E., Bohnsack M.T. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18:2004–2014. doi: 10.15252/embr.201744940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lence T., Paolantoni C., Worpenberg L., Roignant J.Y. Mechanistic insights into m6A RNA enzymes. Biochim. Biophys. Acta. Gene Regul. Mech. 2019;1862:222–229. doi: 10.1016/j.bbagrm.2018.10.014. [DOI] [PubMed] [Google Scholar]
- 15.Das A., Gorospe M., Panda A.C. The coding potential of circRNAs. Aging (Albany N.Y.) 2018;10:2228–2229. doi: 10.18632/aging.101554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou C., Molinie B., Daneshvar K., Pondick J.V., Wang J., Van Wittenberghe N., Xing Y., Giallourakis C.C., Mullen A.C. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 2017;20:2262–2276. doi: 10.1016/j.celrep.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao W., Cui Y., Liu L., Qi X., Liu J., Ma S., Hu X., Zhang Z., Wang Y., Li H. Splicing factor derived circular RNA circUHRF1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. Cell Death Differ. 2019 doi: 10.1038/s41418-019-0423-5. Published online September 30, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuncel G., Kalkan R. Importance of m N6-methyladenosine (m6A) RNA modification in cancer. Med. Oncol. 2019;36:36. doi: 10.1007/s12032-019-1260-6. [DOI] [PubMed] [Google Scholar]
- 19.Chen B., Li Y., Song R., Xue C., Xu F. Functions of RNA N6-methyladenosine modification in cancer progression. Mol. Biol. Rep. 2019;46:2567–2575. doi: 10.1007/s11033-019-04655-4. [DOI] [PubMed] [Google Scholar]
- 20.Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Śledź P., Jinek M. Structural insights into the molecular mechanism of the m6A writer complex. eLife. 2016;5:e18434. doi: 10.7554/eLife.18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang P., Doxtader K.A., Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell. 2016;63:306–317. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J., Dong X., Gong Z., Qin L.Y., Yang S., Zhu Y.L., Wang X., Zhang D., Zou T., Yin P., Tang C. Solution structure of the RNA recognition domain of METTL3-METTL14 N6-methyladenosine methyltransferase. Protein Cell. 2019;10:272–284. doi: 10.1007/s13238-018-0518-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang H., Weng H., Zhou K., Wu T., Zhao B.S., Sun M., Chen Z., Deng X., Xiao G., Auer F. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature. 2019;567:414–419. doi: 10.1038/s41586-019-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ping X.L., Sun B.F., Wang L., Xiao W., Yang X., Wang W.J., Adhikari S., Shi Y., Lv Y., Chen Y.S. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schöller E., Weichmann F., Treiber T., Ringle S., Treiber N., Flatley A., Feederle R., Bruckmann A., Meister G. Interactions, localization, and phosphorylation of the m6A generating METTL3-METTL14-WTAP complex. RNA. 2018;24:499–512. doi: 10.1261/rna.064063.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yue Y., Liu J., Cui X., Cao J., Luo G., Zhang Z., Cheng T., Gao M., Shu X., Ma H. VIRMA mediates preferential m6A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz S., Mumbach M.R., Jovanovic M., Wang T., Maciag K., Bushkin G.G., Mertins P., Ter-Ovanesyan D., Habib N., Cacchiarelli D. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knuckles P., Lence T., Haussmann I.U., Jacob D., Kreim N., Carl S.H., Masiello I., Hares T., Villaseñor R., Hess D. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32:415–429. doi: 10.1101/gad.309146.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horiuchi K., Kawamura T., Iwanari H., Ohashi R., Naito M., Kodama T., Hamakubo T. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 2013;288:33292–33302. doi: 10.1074/jbc.M113.500397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doxtader K.A., Wang P., Scarborough A.M., Seo D., Conrad N.K., Nam Y. Structural basis for regulation of METTL16, an S-adenosylmethionine homeostasis factor. Mol. Cell. 2018;71:1001–1011.e4. doi: 10.1016/j.molcel.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shima H., Matsumoto M., Ishigami Y., Ebina M., Muto A., Sato Y., Kumagai S., Ochiai K., Suzuki T., Igarashi K. S-adenosylmethionine synthesis is regulated by selective N6-adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell Rep. 2017;21:3354–3363. doi: 10.1016/j.celrep.2017.11.092. [DOI] [PubMed] [Google Scholar]
- 33.Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.M., Li C.J., Vågbø C.B., Shi Y., Wang W.L., Song S.H. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G., He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rau K., Rösner L., Rentmeister A. Sequence-specific m6A demethylation in RNA by FTO fused to RCas9. RNA. 2019;25:1311–1323. doi: 10.1261/rna.070706.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei J., Liu F., Lu Z., Fei Q., Ai Y., He P.C., Shi H., Cui X., Su R., Klungland A. Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell. 2018;71:973–985.e5. doi: 10.1016/j.molcel.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mauer J., Luo X., Blanjoie A., Jiao X., Grozhik A.V., Patil D.P., Linder B., Pickering B.F., Vasseur J.J., Chen Q. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mauer J., Sindelar M., Despic V., Guez T., Hawley B.R., Vasseur J.J., Rentmeister A., Gross S.S., Pellizzoni L., Debart F. FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nat. Chem. Biol. 2019;15:340–347. doi: 10.1038/s41589-019-0231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang C., Klukovich R., Peng H., Wang Z., Yu T., Zhang Y., Zheng H., Klungland A., Yan W. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. USA. 2018;115:E325–E333. doi: 10.1073/pnas.1717794115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zou S., Toh J.D., Wong K.H., Gao Y.G., Hong W., Woon E.C.N. N6-methyladenosine: a conformational marker that regulates the substrate specificity of human demethylases FTO and ALKBH5. Sci. Rep. 2016;6:25677. doi: 10.1038/srep25677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C. N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roundtree I.A., Luo G.Z., Zhang Z., Wang X., Zhou T., Cui Y., Sha J., Huang X., Guerrero L., Xie P. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife. 2017;6:e31311. doi: 10.7554/eLife.31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alarcón C.R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S.F. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi H., Wang X., Lu Z., Zhao B.S., Ma H., Hsu P.J., Liu C., He C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsu P.J., Zhu Y., Ma H., Guo Y., Shi X., Liu Y., Qi M., Lu Z., Shi H., Wang J. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115–1127. doi: 10.1038/cr.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wojtas M.N., Pandey R.R., Mendel M., Homolka D., Sachidanandam R., Pillai R.S. Regulation of m6A transcripts by the 3′→5′ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol. Cell. 2017;68:374–387.e12. doi: 10.1016/j.molcel.2017.09.021. [DOI] [PubMed] [Google Scholar]
- 48.Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., Zhao B.S., Mesquita A., Liu C., Yuan C.L. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vu L.P., Pickering B.F., Cheng Y., Zaccara S., Nguyen D., Minuesa G., Chou T., Chow A., Saletore Y., MacKay M. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017;23:1369–1376. doi: 10.1038/nm.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barbieri I., Tzelepis K., Pandolfini L., Shi J., Millán-Zambrano G., Robson S.C., Aspris D., Migliori V., Bannister A.J., Han N. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature. 2017;552:126–131. doi: 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choe J., Lin S., Zhang W., Liu Q., Wang L., Ramirez-Moya J., Du P., Kim W., Tang S., Sliz P. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–560. doi: 10.1038/s41586-018-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weng H., Huang H., Wu H., Qin X., Zhao B.S., Dong L., Shi H., Skibbe J., Shen C., Hu C. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell. 2018;22:191–205.e9. doi: 10.1016/j.stem.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pendleton K.E., Chen B., Liu K., Hunter O.V., Xie Y., Tu B.P., Conrad N.K. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169:824–835.e14. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bansal H., Yihua Q., Iyer S.P., Ganapathy S., Proia D.A., Penalva L.O., Uren P.J., Suresh U., Carew J.S., Karnad A.B. WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia. 2014;28:1171–1174. doi: 10.1038/leu.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sorci M., Ianniello Z., Cruciani S., Larivera S., Ginistrelli L.C., Capuano E., Marchioni M., Fazi F., Fatica A. METTL3 regulates WTAP protein homeostasis. Cell Death Dis. 2018;9:796. doi: 10.1038/s41419-018-0843-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takeda A., Shimada A., Hamamoto K., Yoshino S., Nagai T., Fujii Y., Yamada M., Nakamura Y., Watanabe T., Watanabe Y. Detection of RBM15-MKL1 fusion was useful for diagnosis and monitoring of minimal residual disease in infant acute megakaryoblastic leukemia. Acta Med. Okayama. 2014;68:119–123. doi: 10.18926/AMO/52408. [DOI] [PubMed] [Google Scholar]
- 57.Ma Z., Morris S.W., Valentine V., Li M., Herbrick J.A., Cui X., Bouman D., Li Y., Mehta P.K., Nizetic D. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat. Genet. 2001;28:220–221. doi: 10.1038/90054. [DOI] [PubMed] [Google Scholar]
- 58.Lee J.H., Skalnik D.G. Rbm15-Mkl1 interacts with the Setd1b histone H3-Lys4 methyltransferase via a SPOC domain that is required for cytokine-independent proliferation. PLoS One. 2012;7:e42965. doi: 10.1371/journal.pone.0042965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma X., Renda M.J., Wang L., Cheng E.C., Niu C., Morris S.W., Chi A.S., Krause D.S. Rbm15 modulates Notch-induced transcriptional activation and affects myeloid differentiation. Mol. Cell. Biol. 2007;27:3056–3064. doi: 10.1128/MCB.01339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Niu C., Zhang J., Breslin P., Onciu M., Ma Z., Morris S.W. c-Myc is a target of RNA-binding motif protein 15 in the regulation of adult hematopoietic stem cell and megakaryocyte development. Blood. 2009;114:2087–2096. doi: 10.1182/blood-2009-01-197921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L., Tran N.T., Su H., Wang R., Lu Y., Tang H., Aoyagi S., Guo A., Khodadadi-Jamayran A., Zhou D. Cross-talk between PRMT1-mediated methylation and ubiquitylation on RBM15 controls RNA splicing. eLife. 2015;4:e07938. doi: 10.7554/eLife.07938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jin S., Mi Y., Song J., Zhang P., Liu Y. PRMT1-RBM15 axis regulates megakaryocytic differentiation of human umbilical cord blood CD34+ cells. Exp. Ther. Med. 2018;15:2563–2568. doi: 10.3892/etm.2018.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Z., Weng H., Su R., Weng X., Zuo Z., Li C., Huang H., Nachtergaele S., Dong L., Hu C. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Akichika S., Hirano S., Shichino Y., Suzuki T., Nishimasu H., Ishitani R., Sugita A., Hirose Y., Iwasaki S., Nureki O., Suzuki T. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science. 2019;363:eaav0080. doi: 10.1126/science.aav0080. [DOI] [PubMed] [Google Scholar]
- 65.Sun H., Zhang M., Li K., Bai D., Yi C. Cap-specific, terminal N6-methylation by a mammalian m6Am methyltransferase. Cell Res. 2019;29:80–82. doi: 10.1038/s41422-018-0117-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kwok C.T., Marshall A.D., Rasko J.E., Wong J.J. Genetic alterations of m6A regulators predict poorer survival in acute myeloid leukemia. J. Hematol. Oncol. 2017;10:39. doi: 10.1186/s13045-017-0410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Rooij J.D., Masetti R., van den Heuvel-Eibrink M.M., Cayuela J.M., Trka J., Reinhardt D., Rasche M., Sonneveld E., Alonzo T.A., Fornerod M. Recurrent abnormalities can be used for risk group stratification in pediatric AMKL: a retrospective intergroup study. Blood. 2016;127:3424–3430. doi: 10.1182/blood-2016-01-695551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nishizawa Y., Konno M., Asai A., Koseki J., Kawamoto K., Miyoshi N., Takahashi H., Nishida N., Haraguchi N., Sakai D. Oncogene c-Myc promotes epitranscriptome m6A reader YTHDF1 expression in colorectal cancer. Oncotarget. 2017;9:7476–7486. doi: 10.18632/oncotarget.23554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bai Y., Yang C., Wu R., Huang L., Song S., Li W., Yan P., Lin C., Li D., Zhang Y. YTHDF1 regulates tumorigenicity and cancer stem cell-like activity in human colorectal carcinoma. Front. Oncol. 2019;9:332. doi: 10.3389/fonc.2019.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li T., Hu P.S., Zuo Z., Lin J.F., Li X., Wu Q.N., Chen Z.H., Zeng Z.L., Wang F., Zheng J. METTL3 facilitates tumor progression via an m6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol. Cancer. 2019;18:112. doi: 10.1186/s12943-019-1038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu Y., Yang X., Chen Z., Tian L., Jiang G., Chen F., Li J., An P., Lu L., Luo N. m6A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol. Cancer. 2019;18:87. doi: 10.1186/s12943-019-1014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin S., Liu J., Jiang W., Wang P., Sun C., Wang X., Chen Y., Wang H. METTL3 promotes the proliferation and mobility of gastric cancer cells. Open Med. (Wars.) 2019;14:25–31. doi: 10.1515/med-2019-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu T., Yang S., Sui J., Xu S.Y., Cheng Y.P., Shen B., Zhang Y., Zhang X.M., Yin L.H., Pu Y.P., Liang G.Y. Dysregulated N6-methyladenosine methylation writer METTL3 contributes to the proliferation and migration of gastric cancer. J. Cell. Physiol. 2020;235:548–562. doi: 10.1002/jcp.28994. [DOI] [PubMed] [Google Scholar]
- 74.Zhang C., Zhang M., Ge S., Huang W., Lin X., Gao J., Gong J., Shen L. Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer Med. 2019;8:4766–4781. doi: 10.1002/cam4.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu D., Shao W., Jiang Y., Wang X., Liu Y., Liu X. FTO expression is associated with the occurrence of gastric cancer and prognosis. Oncol. Rep. 2017;38:2285–2292. doi: 10.3892/or.2017.5904. [DOI] [PubMed] [Google Scholar]
- 76.Wu L., Wu D., Ning J., Liu W., Zhang D. Changes of N6-methyladenosine modulators promote breast cancer progression. BMC Cancer. 2019;19:326. doi: 10.1186/s12885-019-5538-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ni T.K., Elman J.S., Jin D.X., Gupta P.B., Kuperwasser C. Premature polyadenylation of MAGI3 is associated with diminished N6-methyladenosine in its large internal exon. Sci. Rep. 2018;8:1415. doi: 10.1038/s41598-018-19916-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Niu Y., Lin Z., Wan A., Chen H., Liang H., Sun L., Wang Y., Li X., Xiong X.F., Wei B. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol. Cancer. 2019;18:46. doi: 10.1186/s12943-019-1004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Klinge C.M., Piell K.M., Tooley C.S., Rouchka E.C. HNRNPA2/B1 is upregulated in endocrine-resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF-7 cells. Sci. Rep. 2019;9:9430. doi: 10.1038/s41598-019-45636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fry N.J., Law B.A., Ilkayeva O.R., Carraway K.R., Holley C.L., Mansfield K.D.N. N6-methyladenosine contributes to cellular phenotype in a genetically-defined model of breast cancer progression. Oncotarget. 2018;9:31231–31243. doi: 10.18632/oncotarget.25782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li J., Han Y., Zhang H., Qian Z., Jia W., Gao Y., Zheng H., Li B. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem. Biophys. Res. Commun. 2019;512:479–485. doi: 10.1016/j.bbrc.2019.03.093. [DOI] [PubMed] [Google Scholar]
- 82.Liu J., Ren D., Du Z., Wang H., Zhang H., Jin Y. m6A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem. Biophys. Res. Commun. 2018;502:456–464. doi: 10.1016/j.bbrc.2018.05.175. [DOI] [PubMed] [Google Scholar]
- 83.Wei W., Huo B., Shi X. miR-600 inhibits lung cancer via downregulating the expression of METTL3. Cancer Manag. Res. 2019;11:1177–1187. doi: 10.2147/CMAR.S181058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brown J.A., Kinzig C.G., DeGregorio S.J., Steitz J.A. Methyltransferase-like protein 16 binds the 3′-terminal triple helix of MALAT1 long noncoding RNA. Proc. Natl. Acad. Sci. USA. 2016;113:14013–14018. doi: 10.1073/pnas.1614759113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo J., Wu Y., Du J., Yang L., Chen W., Gong K., Dai J., Miao S., Jin D., Xi S. Deregulation of UBE2C-mediated autophagy repression aggravates NSCLC progression. Oncogenesis. 2018;7:49. doi: 10.1038/s41389-018-0054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen M., Wei L., Law C.T., Tsang F.H., Shen J., Cheng C.L., Tsang L.H., Ho D.W., Chiu D.K., Lee J.M. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 87.Zhao X., Chen Y., Mao Q., Jiang X., Jiang W., Chen J., Xu W., Zhong L., Sun X. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. 2018;21:859–868. doi: 10.3233/CBM-170791. [DOI] [PubMed] [Google Scholar]
- 88.Zhou Y., Yin Z., Hou B., Yu M., Chen R., Jin H., Jian Z. Expression profiles and prognostic significance of RNA N6-methyladenosine-related genes in patients with hepatocellular carcinoma: evidence from independent datasets. Cancer Manag. Res. 2019;11:3921–3931. doi: 10.2147/CMAR.S191565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ma J.Z., Yang F., Zhou C.C., Liu F., Yuan J.H., Wang F., Wang T.T., Xu Q.G., Zhou W.P., Sun S.H. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6-methyladenosine-dependent primary microRNA processing. Hepatology. 2017;65:529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 90.Cheng X., Li M., Rao X., Zhang W., Li X., Wang L., Huang G. KIAA1429 regulates the migration and invasion of hepatocellular carcinoma by altering m6A modification of ID2 mRNA. OncoTargets Ther. 2019;12:3421–3428. doi: 10.2147/OTT.S180954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang S., Wei J., Cui Y.H., Park G., Shah P., Deng Y., Aplin A.E., Lu Z., Hwang S., He C., He Y.Y. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 2019;10:2782. doi: 10.1038/s41467-019-10669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dahal U., Le K., Gupta M. RNA m6A methyltransferase METTL3 regulates invasiveness of melanoma cells by matrix metallopeptidase 2. Melanoma Res. 2019;29:382–389. doi: 10.1097/CMR.0000000000000580. [DOI] [PubMed] [Google Scholar]
- 93.Cui Q., Shi H., Ye P., Li L., Qu Q., Sun G., Sun G., Lu Z., Huang Y., Yang C.G. m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang S., Zhao B.S., Zhou A., Lin K., Zheng S., Lu Z., Chen Y., Sulman E.P., Xie K., Bögler O. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606.e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Visvanathan A., Patil V., Abdulla S., Hoheisel J.D., Somasundaram K. N6-methyladenosine landscape of glioma stem-like cells: METTL3 is essential for the expression of actively transcribed genes and sustenance of the oncogenic signaling. Genes (Basel) 2019;10:E141. doi: 10.3390/genes10020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Taketo K., Konno M., Asai A., Koseki J., Toratani M., Satoh T., Doki Y., Mori M., Ishii H., Ogawa K. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int. J. Oncol. 2018;52:621–629. doi: 10.3892/ijo.2017.4219. [DOI] [PubMed] [Google Scholar]
- 97.Yang F., Jin H., Que B., Chao Y., Zhang H., Ying X., Zhou Z., Yuan Z., Su J., Wu B. Dynamic m6A mRNA methylation reveals the role of METTL3-m6A-CDCP1 signaling axis in chemical carcinogenesis. Oncogene. 2019;38:4755–4772. doi: 10.1038/s41388-019-0755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Uddin M.B., Roy K.R., Hosain S.B., Khiste S.K., Hill R.A., Jois S.D., Zhao Y., Tackett A.J., Liu Y.Y. An N6-methyladenosine at the transited codon 273 of p53 pre-mRNA promotes the expression of R273H mutant protein and drug resistance of cancer cells. Biochem. Pharmacol. 2019;160:134–145. doi: 10.1016/j.bcp.2018.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu J., Eckert M.A., Harada B.T., Liu S.M., Lu Z., Yu K., Tienda S.M., Chryplewicz A., Zhu A.C., Yang Y. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018;20:1074–1083. doi: 10.1038/s41556-018-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huang Y., Su R., Sheng Y., Dong L., Dong Z., Xu H., Ni T., Zhang Z.S., Zhang T., Li C. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. 2019;35:677–691.e10. doi: 10.1016/j.ccell.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Su R.D.L., Li C., Nachtergaele S., Wunderlich M., Qing Y., Deng X., Wang Y., Weng X., Hu C. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell 2017;172:90–105.e23. R-2HG Targets FTO to Increase m(6)A Levels and Suppress Tumor Growth. Cancer Discov. 2018;8:137. [Google Scholar]
- 102.Zhou S., Bai Z.L., Xia D., Zhao Z.J., Zhao R., Wang Y.Y., Zhe H. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol. Carcinog. 2018;57:590–597. doi: 10.1002/mc.22782. [DOI] [PubMed] [Google Scholar]
- 103.Zhuang C., Zhuang C., Luo X., Huang X., Yao L., Li J., Li Y., Xiong T., Ye J., Zhang F., Gui Y. N6-methyladenosine demethylase FTO suppresses clear cell renal cell carcinoma through a novel FTO-PGC-1α signalling axis. J. Cell. Mol. Med. 2019;23:2163–2173. doi: 10.1111/jcmm.14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li Y., Zheng D., Wang F., Xu Y., Yu H., Zhang H. Expression of demethylase genes, FTO and ALKBH1, is associated with prognosis of gastric cancer. Dig. Dis. Sci. 2019;64:1503–1513. doi: 10.1007/s10620-018-5452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lin X., Chai G., Wu Y., Li J., Chen F., Liu J., Luo G., Tauler J., Du J., Lin S. RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat. Commun. 2019;10:2065. doi: 10.1038/s41467-019-09865-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 106.Han D., Liu J., Chen C., Dong L., Liu Y., Chang R., Huang X., Liu Y., Wang J., Dougherty U. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature. 2019;566:270–274. doi: 10.1038/s41586-019-0916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Paris J., Morgan M., Campos J., Spencer G.J., Shmakova A., Ivanova I., Mapperley C., Lawson H., Wotherspoon D.A., Sepulveda C. Targeting the RNA m6A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell. 2019;25:137–148.e6. doi: 10.1016/j.stem.2019.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li Z., Qian P., Shao W., Shi H., He X.C., Gogol M., Yu Z., Wang Y., Qi M., Zhu Y. Suppression of m6A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 2018;28:904–917. doi: 10.1038/s41422-018-0072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]