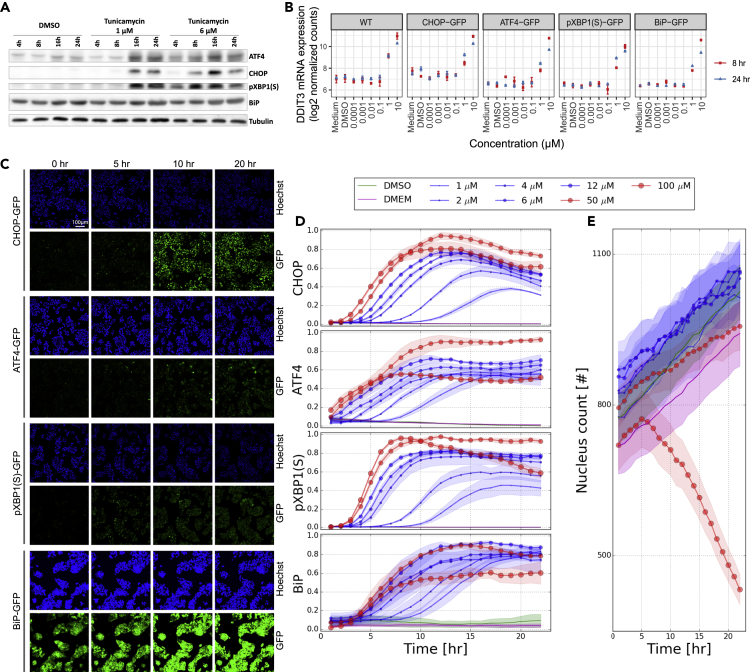
Figure 2.
Dynamic Measurements of Various UPR Components to Integrate with Modeling
(A) Western blot of CHOP, ATF4, pXBP1(S), and BiP protein at 4, 8, 16, and 24 h upon exposure to DMSO or tunicamycin (1 and 6 μM) in WT HepG2 cells. Tubulin was used as protein loading control.
(B) Log2 normalized counts of DDIT3 mRNA expression analyzed using TempO-seq transcriptomics at 8 or 24 h after exposure with various concentrations of tunicamycin in HepG2 WT and UPR BAC-GFP reporter cell lines.
(C) Representative images of HepG2 UPR BAC-GFP reporter cell lines (CHOP, ATF4, pXBP1(S), and BiP) stained with Hoechst for nuclei visualization. Images were obtained using confocal microscopy with a 20× objective at the indicated time points after exposure to tunicamycin at 6 μM. Hoechst is represented in blue (upper rows) and GFP in green (lower rows).
(D and E) Quantification of single-cell-based GFP intensity of the HepG2 UPR BAC-GFP reporter cell lines after min-max normalization (D) and cell counts (E) after exposure to DMEM/DMSO or to a broad concentration range of tunicamycin and imaged live every hour for 24 h after exposure using confocal microscopy. BiP-GFP intensity was quantified in the cytoplasm; all other reporters were quantified in the nuclei.
Data in (B), (D), and (E) represent mean and standard error of the mean (SE) of three biological replicates.