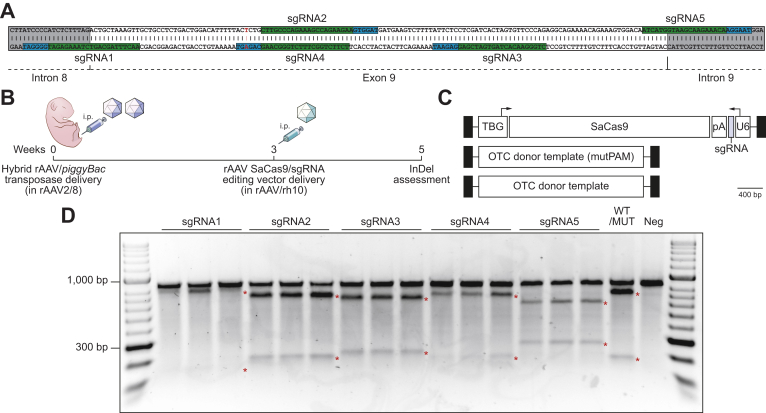
Fig. 2.
Functional analysis of rAAV editing reagents.
(A) The location of sgRNA and SaCas9 PAM sequences are indicated (highlighted in green and blue, respectively). Intronic sequences are shaded in grey and the patient-specific c.905A>T mutation is indicated by the nucleotide shown in red. (B) Overview and timing of intraperitoneal rAAV delivery for CRISPR-SaCas9-mediated gene disruption in Spfash mice. Newborn animals (n = 5 per treatment group) received 5×1010 vg transposase and 1×1011 vg minigene vector (packaged in the AAV8 capsid) and 3 weeks later, 2×1011 of a SaCas9 with sgRNA (1 to 5) rAAV vector (packaged in the rh10 capsid). (C) Configuration of the dual rAAV genome editing vectors used in this study. (D) Functional analysis of sgRNAs used to disrupt the human OTC locus in newborn Spfash mice (n = 5 per treatment group). The presence of small InDels was confirmed by using Surveyor® nuclease. An equal mix of PCR products from WT and mutant minigenes (WT/MUT) and mutant only (neg) were included as controls for the Surveyor® reaction. Red asterisks indicate expected cleavage fragments. InDels, insertions and deletions; pA, bovine growth hormone polyadenylation signal sequence; PAM, protospacer adjacent motif; rAAV, recombinant adeno-associated virus; SaCas9, Staphylococcus aureus Cas9 nuclease; sgRNA, single guide RNA; TBG, human thyroxine binding globulin promoter; U6, RNA polymerase III promoter for human U6 snRNA; WT, wild-type.