Figure 1.
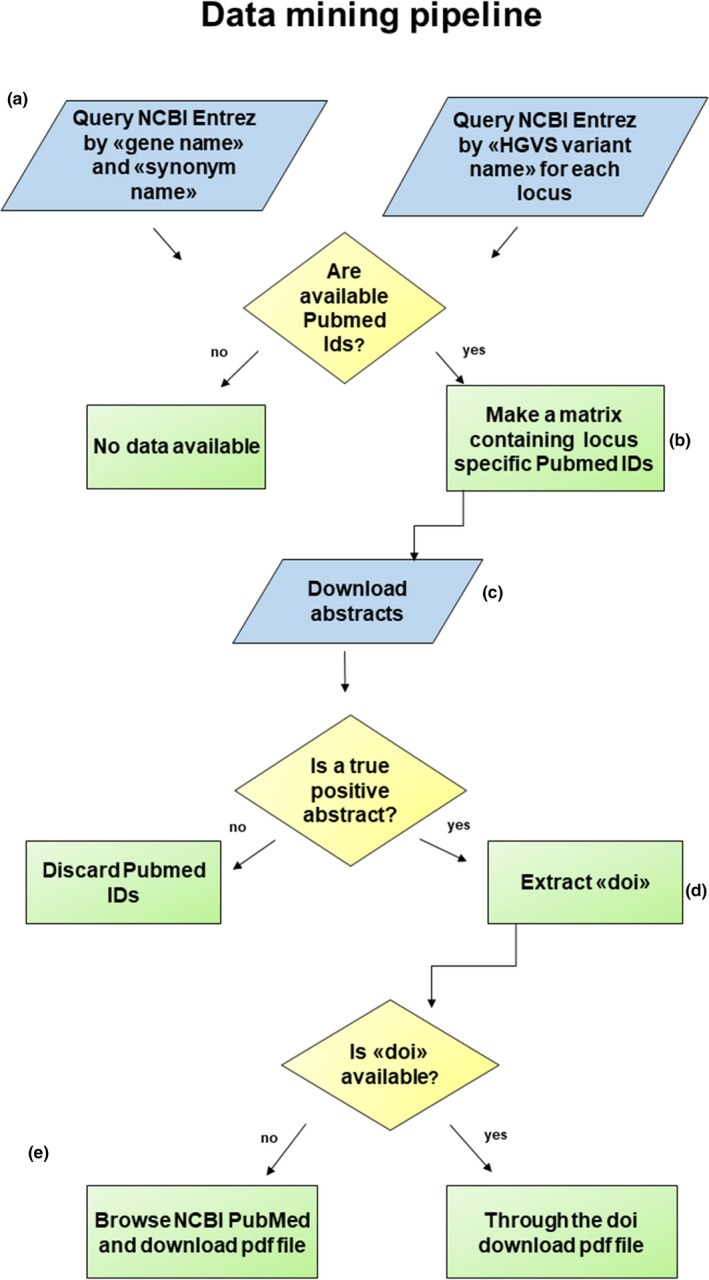
Workflow describing data mining pipeline. For each mtDNA locus, the main steps are: (a) query through the NCBI Entrez system the PubMed database by “gene name or synonym gene name” and “mtDNA variant name” in HGVS format; (b) store the retrieved Pubmed IDs list; (c) download the abstract related to each Pubmed IDs and keep those containing information regarding functional studies and/or variant; (d) for true positive Pubmed IDs, extract the DOI; e) browse NCBI PubMed and download the related PDF articles
