Figure 7.
Creation of an RsiG Derivative Defective in c-di-GMP Binding (RsiG∗) and Demonstration that c-di-GMP Binding Is Required for RsiG Activity In Vivo
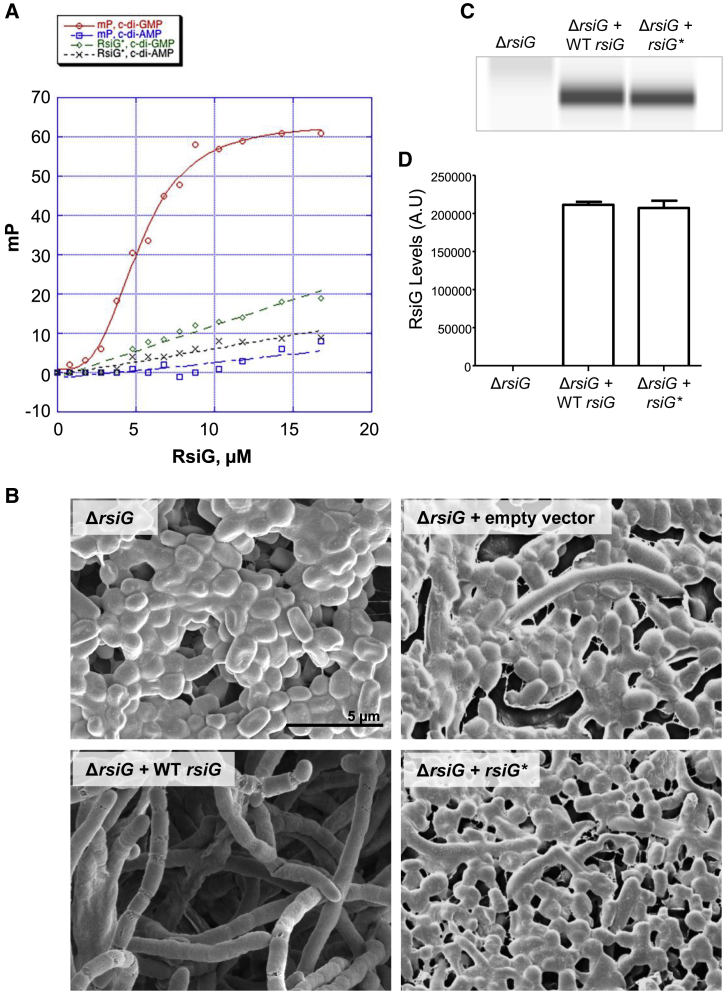
(A) Fluorescence polarization (FP)-based binding isotherms of WT RsiG binding to 2′-Fluo-AHC-c-di-GMP (red circles), WT RsiG binding to 2′-Fluo-AHC-c-di-AMP (blue squares), RsiG∗ binding to 2′-Fluo-AHC-c-di-GMP (green diamonds), and RsiG∗ binding to 2′-Fluo-AHC-c-di-AMP (black crosses). Each binding curve is a representative from 3 technical replicates. The y axis shows millipolarization units (mP), and the x axis shows the concentration of RsiG protein (micromolar).
(B) Scanning electron micrographs showing that deletion of rsiG causes hypersporulation and that the rsiG mutant can be complemented with WT rsiG but that it cannot be complemented with the rsiG∗ allele (R71A, D79A, R169A, D177A) encoding a protein defective in c-di-GMP binding. Strains were imaged after 7 days of growth on DNA medium.
(C) RsiG∗ is not affected regarding protein stability. Shown is an automated western blot analysis comparing RsiG protein levels in the rsiG mutant complemented with the WT rsiG allele and the rsiG∗ allele, generated using the quantitative Wes capillary electrophoresis and blotting system (ProteinSimple, San Jose, CA; STAR Methods). Equal amounts (2.5 μg) of total protein were loaded for each sample, and RsiG was detected with a polyclonal anti-RsiG antibody. A single replicate is shown for each strain. Strains were grown in DNB, and samples were collected at 15 h.
(D) Quantification of RsiG levels (area under each peak, a.u.).
All samples were analyzed in triplicate, and the mean value and its SE are shown for each sample. See also Figure S4 for biochemical analysis of the dependence of complex formation on c-di-GMP as well as Figure S5 for conservation of c-di-GMP-binding residues among homologs of RsiG and σWhiG.