Figure 1.
High-Resolution Proteome of the Somatosensory Cortex and Neurogenic Niches
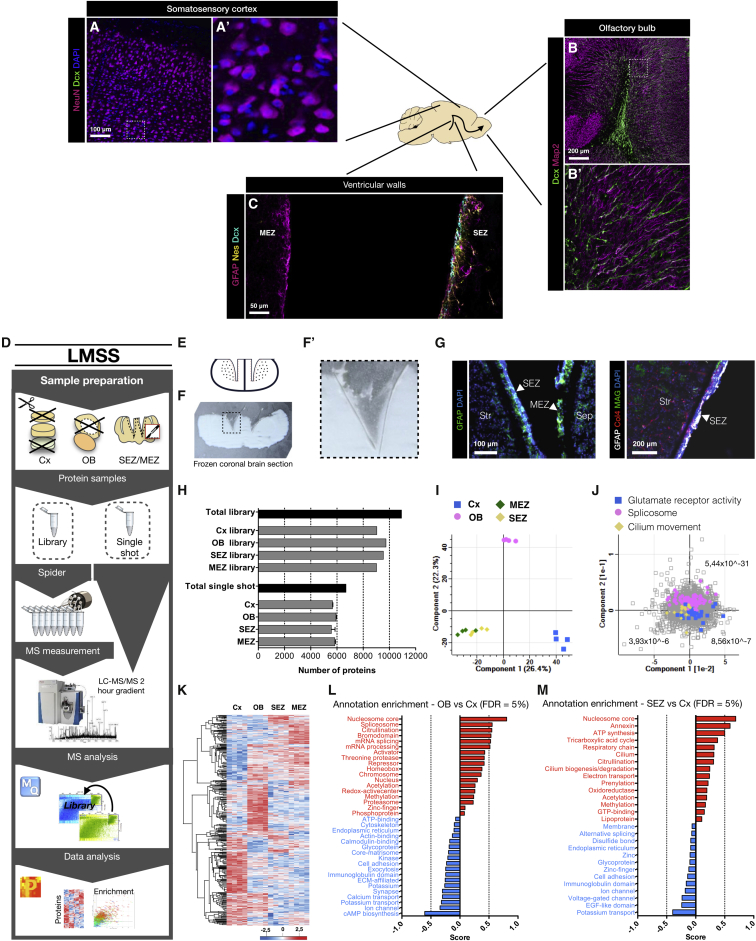
(A–C) The schematic drawing indicates a sagittal section of the adult murine brain with example photomicrographs of the regions used in this analysis—the non-neurogenic somatosensory cortex (A), the olfactory bulb (OB), where new neurons (labeled for doublecortin [Dcx]) integrate (B), and the lateral sub-ependymal zone (SEZ) where most NSCs reside, whereas only a few are located in the medial sub-ependymal zone (MEZ) (C). Sections were immunostained as indicated in the panels and are confocal z stacks.
(D) Experimental workflow using loss-less nano-fractionation for library-matched single shot measurements.
(E) Schematic of the high-precision cryo-dissection of the SEZ and the MEZ.
(F) Picture of a 50-μm frozen coronal section (white, ventral down) with cortex, corpus callosum, and choroid plexus removed. (F′) shows magnification of the dissected region visible as a thin gray line.
(G) Photomicrographs of cryo-dissected SEZ and MEZ (separated from striatum [Str] and septum [Sep]) stained for GFAP and DAPI (left panel) and cryo-dissected SEZ stained for GFAP, collagen 4 (Col4), myelin-associated glycoprotein (MAG), and DAPI (right panel).
(H) Number of proteins quantified in the library sample measurements and the library-matched single shot (LMSS) sample measurements for each region. Data are shown as mean ± standard deviation (n = 1 library sample per region, n = 4 single shot samples per region). See also Figures S1A–S1D.
(I) Principal component analysis (PCA) for each brain region. Components 1 and 2 separate the main regions. The SEZ and the MEZ are similar in these components.
(J) Colors indicate three categories that are enriched, respectively, in the Cx, the OB, and both the SEZ and the MEZ (FDR is presented for each category).
(K) Heatmap of 4,786 proteins found to be of different abundance comparing the four brain regions (n = 4 per region). Intensities are based on label-free quantification (LFQ) intensities after unsupervised hierarchical clustering (ANOVA with Benjamin-Hochberg post hoc test, FDR = 0.05).
(L) The datasets were annotated with Uniprot keywords and the matrisome annotation (see STAR Methods). Enriched features of the OB in comparison to the Cx were then scored (0 to 1) and are displayed in a bar graph (1D-annotation enrichment, FDR = 0.05). Conversely, features with a negative score (0 to −1) are enriched in the Cx compared to the OB.
(M) Enriched features of the SEZ in comparison to the Cx were analyzed in the same manner (1D-annotation enrichment, FDR = 0.05).
Scale bars as indicated in the panels.