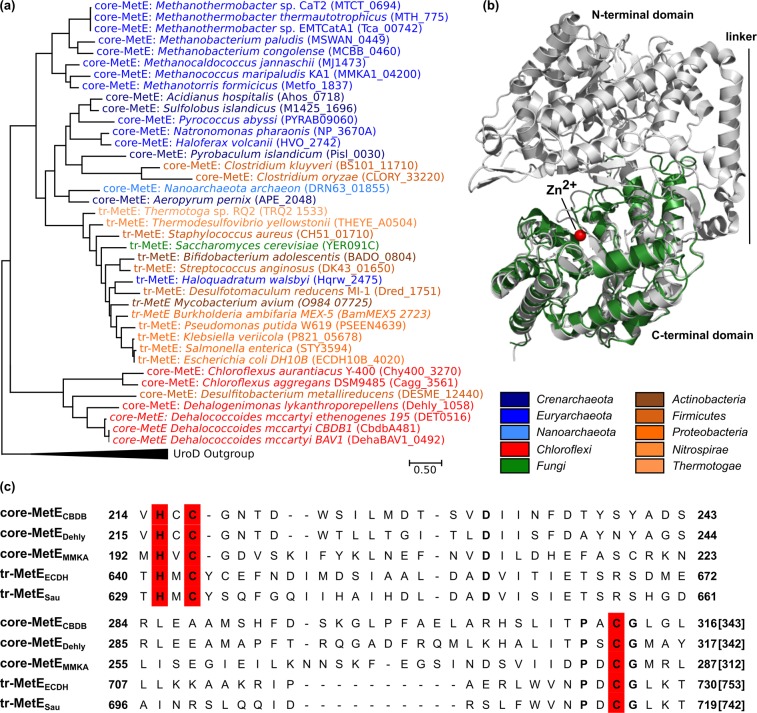
Figure 1.
Bioinformatic analysis of the core-MetECBDB from Dehalococcoides mccartyi strain CBDB1. (a) Maximum-Likelihood phylogenetic tree of MetE representatives was generated with MEGA767. Multiple amino acid sequence alignments of full length with deletion of gaps (MUSCLE algorithm) were used to generate the tree. The analysis involved 39 amino acid sequences including the C-terminus of tandem-repeat methionine synthases (tr-MetE) from bacteria (brown colors) and yeast (green) as well as core-MetEs from archaea (blue colors), Chloroflexi (red) and Clostridiales (brown). The gene loci are in brackets. (b) The crystal structure of core-MetECBDB was calculated with the I-TASSER server65. Overlay of the crystal structure of tr-MetE from Neurospora crassa (PDB No. 4ZTX, grey) with the structural model obtained for core-MetECBDB from D. mccartyi strain CBDB1 (green) was obtained with PyMOL66. Core-MetECBDB matches the C-terminal part of tr-MetENcra (C-score = −0.27) but lacks the N-terminal part and the linker region. (c) Amino acid sequence alignment of selected MetE proteins. The Zn2+-binding site HXCXnC (red) is conserved in annotated tr-MetEs and also in core-MetE homologs. Core-MetECBDB: core-MetE from D. mccartyi strain CBDB1, coreMetEDehly: core-MetE from Dehalogenimonas lykanthroporepellens, core-MetEMMKA: core-MetE from Methanococcus maripaludis, tr-MetEECDH: tandem-repeat MetE from Escherichia coli DH10B, tr-MetESau: tandem-repeat MetE from Staphylococcus aureus.