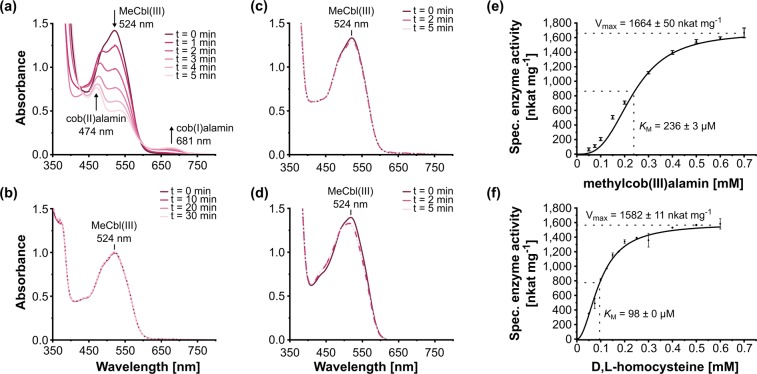
Figure 3.
Demethylation of methylcob(III)alamin (MeCbl(III)) catalyzed by purified core-MetECBDB from D. mccartyi strain CBDB1 in the presence of D,L-homocysteine. (a) In the presence of 0.1 µM core-MetECBDB, 0.5 mM methylcob(III)alamin and 2 mM D,L-homocysteine, continuous changes in the UV/Vis spectrum were observed indicating the consumption of methylcob(III)alamin (524 nm) and the formation of cob(I)alamin (681 nm) and cob(II)alamin (474 nm) as highlighted by arrows. (b) In the absence of core-MetECBDB, the UV/Vis spectrum remained unchanged over an incubation time of 30 min. (c) In the absence of D,L-homocysteine, the UV/Vis spectrum remained unchanged. (d) In the presence of 0.5 mg mL−1 E. coli crude extract, instead of core-MetECBDB as catalyst, MeCbl(III) was not transformed. (e) Dependence of core-MetECBDB methyltransferase activity on methylcob(III)alamin concentration. (f) Dependence of core-MetECBDB methyltransferase activity on D,L-homocysteine concentration. The activities in panels (e,f) were determined by following the change of the absorption at 681 nm. Vmax and KM were calculated according to a Hill-Fit plot with R2 = 0.998 for panel (e) and R2 = 0.999 for panel (f), respectively.