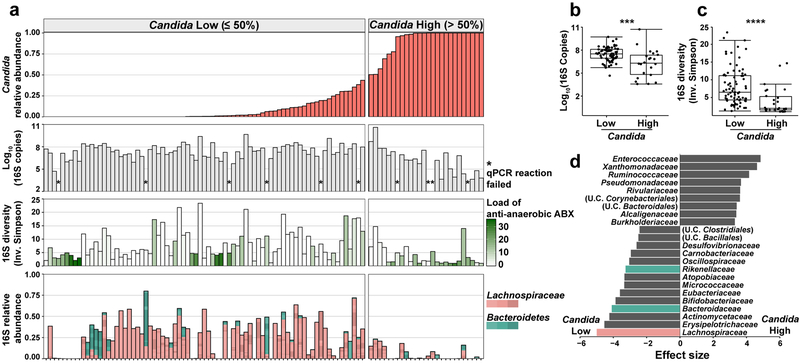
Fig. 4: Characterization of bacteria in fecal samples with high and low levels of pathogenic Candida species.
a, Alignment of fecal samples according to the relative abundance of pathogenic Candida species (1st row), bacterial burden (by 16S rDNA qPCR; 2nd row), bacterial diversity (by Inverse Simpson Index; 3rd row), and relative abundance of Lachnospiraceae and Bacteroidetes (4th row). * indicates failed 16S qPCR reaction. Each bar in the bacterial diversity graph (3rd row) is colored to indicate the anti-anaerobic antibiotic load index prior to sample collection; this index reflects the sum of administered anti-anaerobic antibiotics (metronidazole, clindamycin, carbapenems, piperacillin/tazobactam, and oral vancomycin) multiplied by the number of treatment days40. b, Comparison of bacterial burden in samples with high (> 50%, n = 22) or low (≤ 50%, n = 71) Candida relative abundance. ***: Two-sided Wilcoxon rank sum test p = 0.00073. c, Comparison of bacterial diversity in samples with high (> 50%, n = 26) or low (≤ 50%, n = 77) Candida relative abundance. ****: Two-sided Wilcoxon rank sum test p = 1.9 × 10−5. The box plots in b and c represent the median, interquartile range (IQR) and range. d, LEfSe analysis of bacteria taxa (at family level) present in samples with high (> 50%, n = 26) or low (≤ 50%, n = 77) relative Candida abundance. The effect size corresponds to the linear discriminant analysis (LDA) score. U.C.: unclassified. A subset of the 16S rDNA sequencing data has been previously reported7,20.