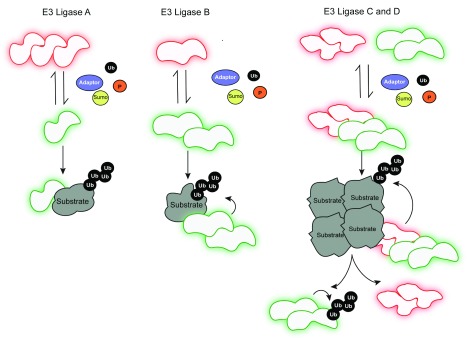
Figure 1. Different types of E3 ligase regulation and assembly.
E3 ligase A is inactive (red) as an oligomer and converted into an active monomer (green) upon post-translational modification or binding to adaptor molecules, indicated with orange, yellow, black, and violet circles, representing phosphate (P), sumo, ubiquitin (Ub), and adaptor molecules, respectively. Conversely, E3 ligase B is inactive as a monomer and activated upon dimerization. Heterotypic interaction of inactive E3 ligase C and active E3 ligase D results in the formation of a multimeric E3 ligase complex, which is able to target oligomeric substrates for ubiquitylation. Upon substrate degradation, the remaining, active ligase D undergoes auto-ubiquitylation and turnover. The different substrates are indicated in other shapes.