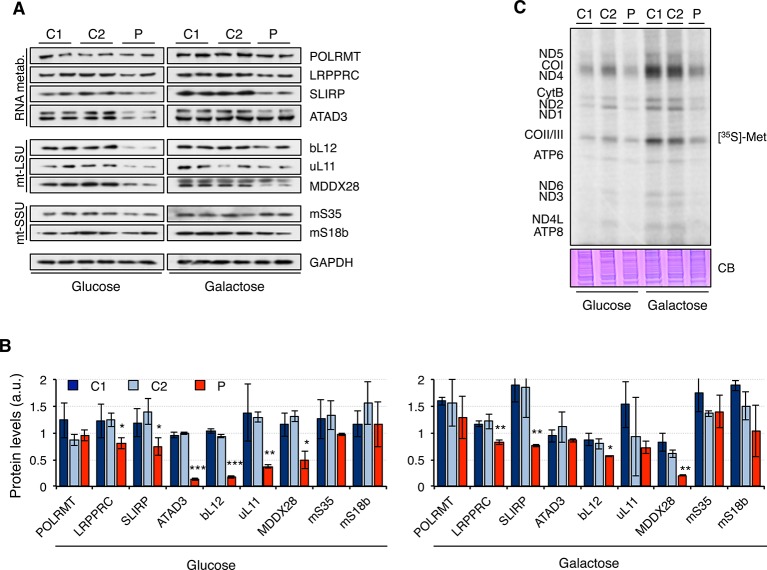
Figure 4.
Mitochondrial translation in mutant RNASEH1 fibroblasts. (A) Western blot analysis of mitochondrial proteins involved in mitochondrial RNA metabolism (RNA metab.) and mitochondrial large (mtLSU) and small (mtSSU) ribosomal subunits in control (C1 and C2) and patient (P) fibroblasts grown in either glucose- or galactose-containing medium. GAPDH was used as loading control. GAPDH is from the same blot as Figure1D . (B) Quantification of the Western blots shown in (A) normalized to GAPDH levels. Data are shown as mean ± SD, Student’s unpaired two-tail t-test, *p < 0.05, **p < 0.01, ***p < 0.001. (C) [35S]-methionine de novo synthesis of mitochondrially encoded proteins in control (C1 and C2) and patient (P) fibroblasts grown in either glucose- or galactose-containing medium. Newly synthesized proteins were visualized after exposure of the dried gel to phosphor screens. The coomassie blue (CB) staining shown below was used as loading control. n = 3.