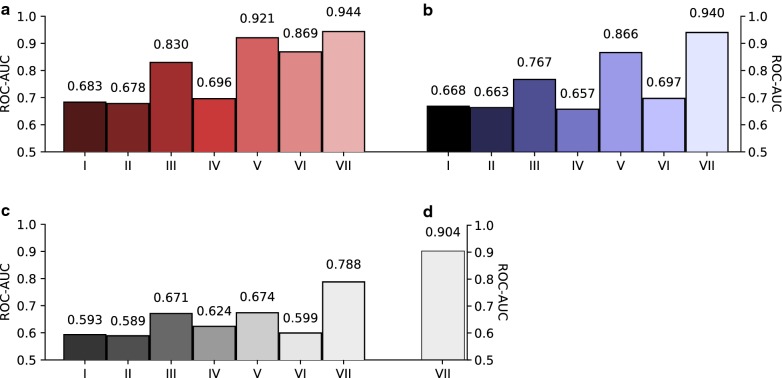
Fig. 2.
Prediction performances of different prioritization tools on test sets, representing various regions of the genome for which the number of features varies. I: whole test set; II: intergenic SNVs; III: transcribed SNVs; IV: SNVs in intron, 5′ and 3′ UTR; V: coding SNVs; VI: SNVs causing synonymous mutations; VII: SNVs causing missense mutations. a pCADD performance measured in ROC-AUC on the different subsets of the pig held-out test set. b mCADD test performance measured in ROC-AUC on the same genomic subsets in the mouse genome. c Performance of 6-taxa laurasiatheria PhastCons conservation score in the pig test set. d SIFT performance on missense causal SNVs in the pig test set