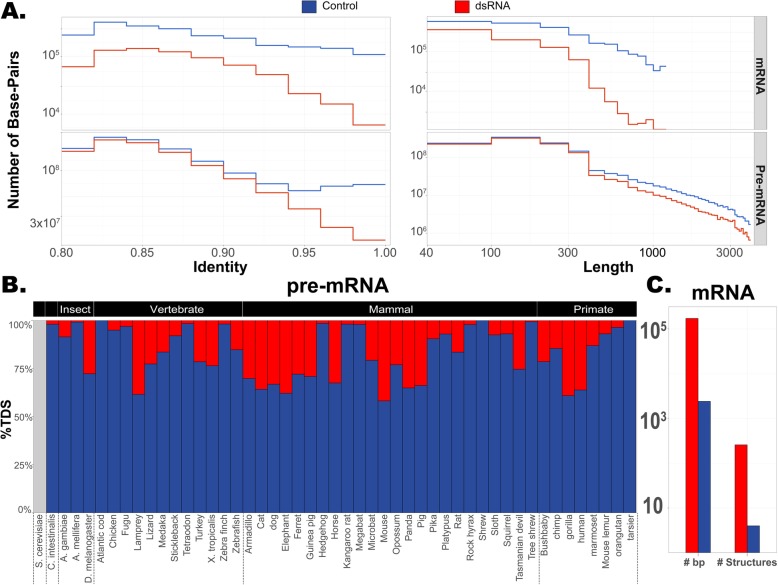
Fig. 3.
Long and nearly perfect duplexes are extremely rare. a Number of genomic nucleotides within putative dsRNAs (IDS) and control (TDS) regions, summed over all organisms, as a function of the region length (right) and the similarity (left), for both mRNA (top) and pre-mRNA (bottom). Note the logarithmic scale. For regions longer than 300 bp and of very high identity, the depletion of putative dsRNAs becomes more pronounced, even in pre-mRNAs. b Comparison of long (> 300), nearly perfect (> 96%), inverted duplicated sequences (potentially folding into dsRNA; red) to tandem duplicated sequences (control; blue) across a wide range of organisms, for pre-mRNA molecules. In most organisms, these putative dsRNA structures are completely depleted. Gray indicates no data (zero IDS and zero TDS). c In mRNA, there are only 4 long and nearly perfect structures in all organisms combined (2419 bps), compared with 258 control TDSs (172,326 bps)