Figure 6.
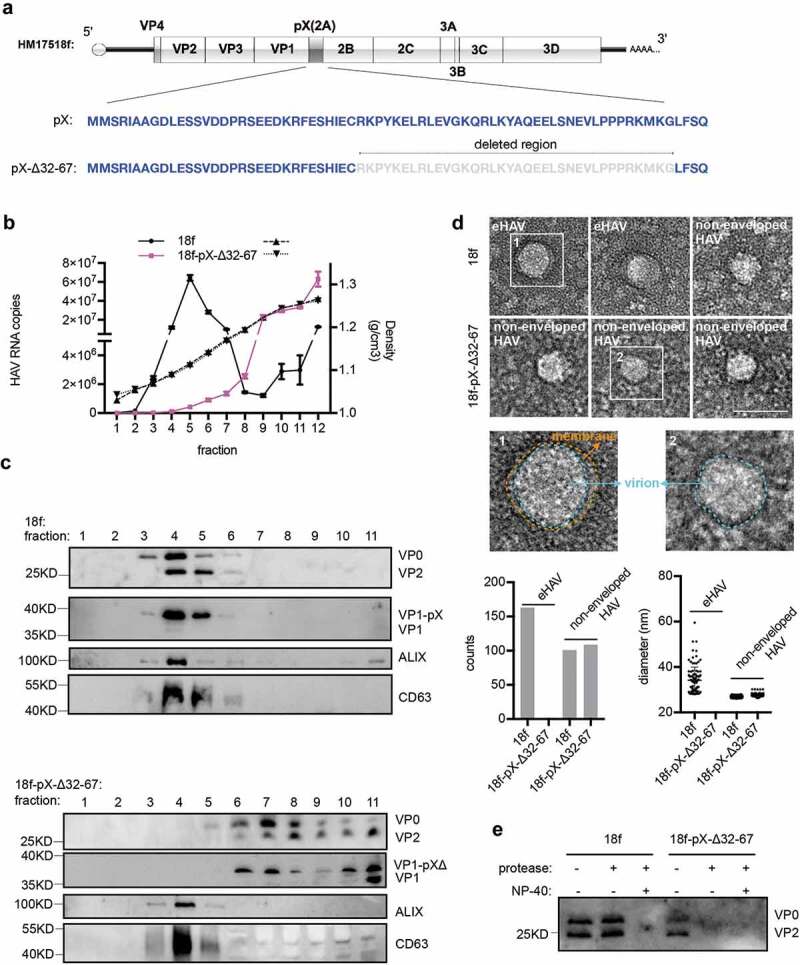
Truncation of the C-terminal 36 aa of pX in the HAV genome abolishes eHAV release. (a) Schematic representation of the HAV genome. Δ indicates truncation. Numbers represent the positions of amino acids. (b) RT-qPCR quantification of HAV genomic RNA in fractions separated by gradient density centrifugation. HM175/18f and HM175/18-pX-Δ32-67 viruses were used to infect Huh7 cells at a multiplicity of infection (MOI) = 1. Four hours after infection, the cells were washed twice with 1× PBS; 1 day postinfection, the medium was replaced by serum-free DMEM. Forty hours after infection, the cell culture supernatant was harvested for concentration. Due to lower virion production from HM175/18-pX-Δ32-67 virus, 20 ml for HM175/18f and 100 ml for HM175/18-pX-Δ32-67 were applied for ultra-filtration and subsequent gradient density centrifugation. (c) Western blot analysis of fractions separated by gradient density centrifugation. Rabbit anti-VP2 was applied to detect extracellular VP0 and VP2. Rabbit anti-VP1 was used to detect VP1 or pX-bound VP1. VP1-pX-Δ represents VP1-pX-Δ32-67. (d) EM analysis of extracellular HAV virions. HAV virus pelleted at 100,000 ×g was resuspended in 1× PBS and prepared for EM. At least 30 pictures in each group from two independent replicates were used for statistical analysis on the numbers of observed virions. In addition, more than 60% of observed structures were subjected to Fiji software for calculation of the diameters of HAV virions. (e) Protease digestion protection assay to validate the naked form of HM175/18-pX-Δ32-67. Viruses resuspended in 1×PBS were treated with the detergent 0.5%NP-40 and protease (50 μg/ml) at 37°C for 5 minutes.
