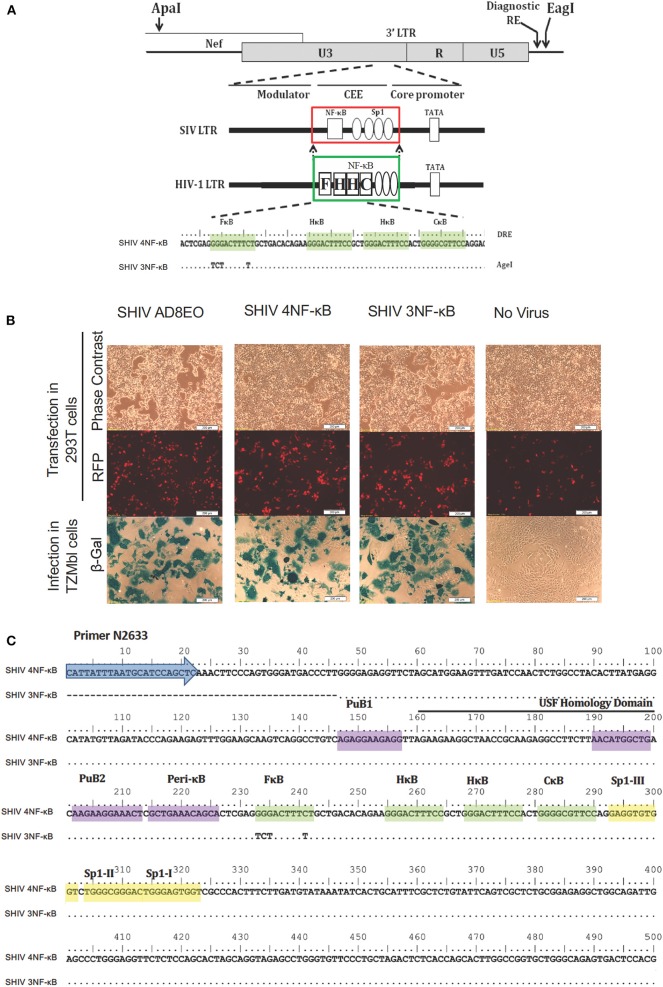
Figure 1.
Construction of promoter-chimera SHIVs with NF-κB duplication. (A) The schematic diagram of 3′ LTR of SIV AD8EO molecular clone. The locations of ApaI and EagI insertion are depicted. The engineered ApaI restriction site does not alter the amino acid sequence in Nef. Each variant strain in the panel contains a diagnostic restriction enzyme (DRE) immediately upstream of EagI for unambiguous identification of the clone. The region of SIV comprising of a single NF-κB binding site (square box) and four Sp1 binding sites (ovals) was substituted with an analogous region of HIV-1C LTR comprising of four NF-κB and three Sp1 binding sites (region substituted is highlighted by square boxes). The parental SHIV promoter-chimera LTR containing four NF-κB binding sites (highlighted by shading) was subsequently replaced by variant LTR in which the 5′ most upstream NF-κB binding site was inactivated. In the sequence alignment, dots represent sequence identity. Base substitutions introduced to inactivate the NF-κB binding sites are shown. (B) The constructs were transfected into 293T. Transfection efficiency was measured by RFP expressed by EF-1α promoter and infectivity of the progeny virions was determined by LTR driven β-galactosidase expression in TZM-bl cells. (C) Sequence confirmation of SHIV AD8EO, SHIV 4NF-κB, and SHIV 3NF-κB molecular clones demonstrates presence of appropriate NF-κB binding sites and other promoter elements.