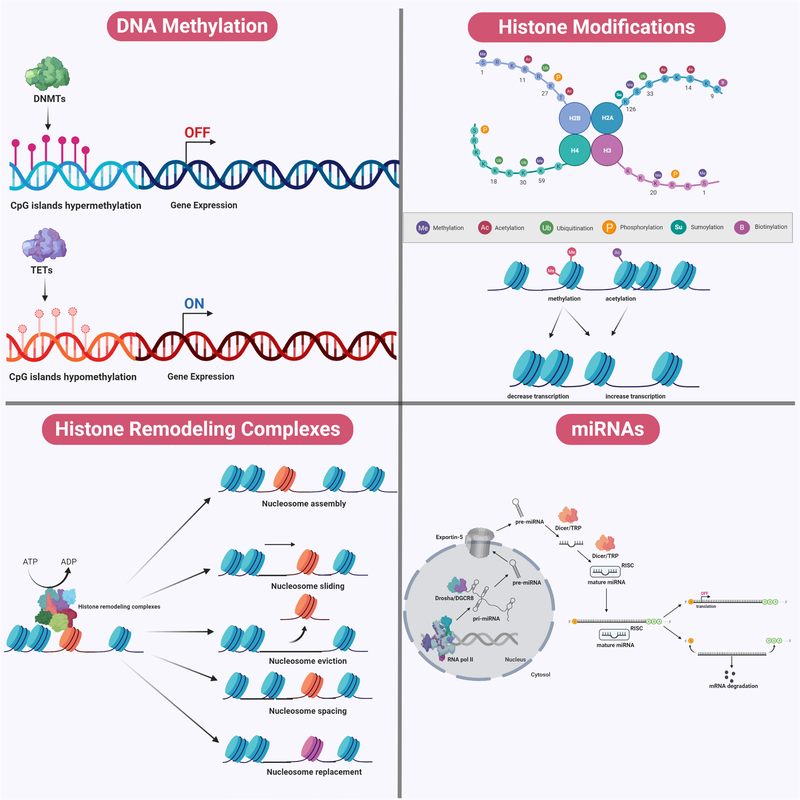
Figure 5.
Mechanisms of Epigenetic Modifications. During DNA methylation, a methyl group is added to carbon position five of the cytosine ring at either CpG dinucleotide islands or CpG shores. DNA methylation is catalyzed by DNMTs, whereas DNA demethylation is catalyzed by TETs. DNA methylation tends to be associated with gene silencing whereas DNA demethylation is associated with transcriptional activation. Chromosomal DNA within the eukaryotic genome is often interlaced with distinct packing and folding, resulting in a distinct organizational structure. At the fundamental level, a nucleosome structural core is first formed through wrapping of a 147 base-pair double helical DNA around an octamer of histone proteins consisting of pairs of H2A-H2B dimers and H3-H4 dimers. Between two nucleosome cores, DNA that is not wrapped around nucleosomes is referred to as ‘linker DNA’, which tends to be associated with H1 histone proteins. A plethora of post-translational modifications (such as acetylation, methylation, phosphorylation, sumoylation, biotinylation, and ubiquitination) can occur on the amino terminal tail protruding from each histone subunit within the nucleosome complex. These histone modifications can affect the relative packing of DNA around these nucleosome cores, and may determine the relative accessibility for the machinery to initiate replication and transcription. Chromatin structure can be modified by ATP-dependent histone remodeling complexes, using the energy from ATP hydrolysis to locally disrupt the interaction between DNA and histones. These remodelers mediate alterations to chromatin structure via nucleosome sliding, eviction, assembly, spacing, histone dimers eviction or replacement, and even entire histone replacement. In addition, miRNAs demonstrate complementary interaction with specific genes and modulate gene expression. Cellular miRNAs are synthesized in a sequential manner and occur in both the nucleus and the cytosol. Within the nucleus, the genome comprises numerous miRNA genes transcribed by RNA polymerase II to form a single large pri-miRNA transcript. Because this pri-miRNAs transcript consists of multiple miRNA loci, it undergoes further processing by a complex called Drosha/DGCR8, a class of RNaseIII enzymatic complex, to generate shorter hairpin-loop structurers termed ‘pre-miRNAs’. This pri-miRNA is exported into the cytosol by Exportin-5 in a RAN-GTP dependent manner. Within the cytosol, another complex belonging to another class of RNaseIII enzymes, termed ‘Dicer/TRBP’ exert its action. Dicer processes the pri-miRNAs into a duplex of mature miRNAs, which undergoes asymmetrical unwinding by the Dicer/TRBP complex to yield a single-stranded mature miRNA. This single-stranded form loads itself into a ribonucleoparticle to yield an RISC, which is the active form of the gene silencing complex. Together, the single-stranded mature miRNAs within the RISC complex will identify the 3’-UTR of targeted mRNAs. The mode of action of gene silencing mediated by miRNAs can occur in two ways, depending on its complementarity with the targeted mRNAs. miRNAs which bind to targeted mRNAs in perfect complementarity induce rapid deadenylation and decapping, leading to mRNA degradation. An imperfect complementarity binding of miRNAs to targeted mRNAs result in translational suppression.
DNMTs, DNA methyltransferase; TETs, ten-eleven translocation; pri-miRNAs, primary miRNA; pre-miRNAs, precursor miRNAs; RISC, RNA-induced silencing complex; 3’-UTR, 3’ untranslated region.