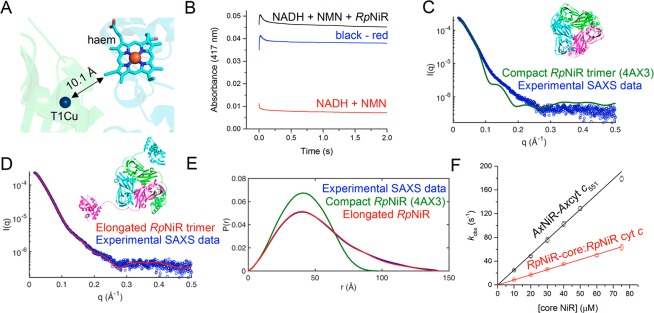
Figure 4.
Domain conformational dynamics limit haem to T1Cu electron transfer in RpNiR. (A) Cartoon representation of the RpNiR structure showing a close-up of the haem and T1Cu centers. (B) Example laser flash photolysis transient for intraprotein electron transfer reaction from the haem to the T1Cu in nitrite-free RpNiR. The red transient shows the background reaction associated with NADH photoexcitation in the presence of the mediator, N-methyl nicotinamide (NMN); the black transient shows the reaction with the RpNiR protein present in the reaction mixture; and the blue transient is the deconvoluted trace, corresponding to the haem to T1Cu electron transfer step in RpNiR. (C) Comparison of experimental small-angle X-ray scattering (SAXS) from full-length wild-type RpNiR33 with that calculated from the crystal structure10 (PDB ID: 4AX3), depicted as inset, χ2 = 21.0. (D) Comparison of experimental SAXS data33 with that calculated for a model, shown as inset, of RpNiR created using the linker conformation of the RpNiR-core structure and refining the position of conformationally plastic cyt c domains, χ2 = 1.5. (E) A comparison of SAXS distance distribution functions (P(R))33 for compact and elongated RpNiR. (F) The rate of interprotein electron transfer from the isolated cytochrome c protein to the core CuNiR proteins.