Table 1. 10 non-redundant transcription factors binding motifs most frequently detected by first layer convolutional filters at FDR < 5%.
Sequence logos of representative CNN filters are shown. Transcription factor binding motifs redundancy was removed with Tomtom motif similarity search with other motifs detected by CNNs with q < 0.05 for similarity to the main motif are listed in the last column; only three motifs with highest similarity are listed.
| Motif name/TF | Representative CNN filter logo |
Motif logo | CNNs with filter match q < 0.05 |
Similar TF motifs discovered |
|---|---|---|---|---|
| M6114_1.02 FOXA1 |
 |
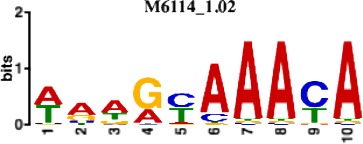 |
838 | M6234_1.02 FOXA3, M6241_1.02 FOXJ2, M4567_1.02 FOXA2… |
| M4427_1.02 CTCF |
 |
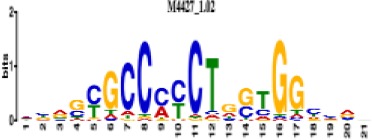 |
833 | M4612_1.02 CTCFL |
| M1906_1.02 SP1 |
 |
 |
677 | M2314_1.02 SP2, M6482_1.02 SP3, M6535_1.02 WT1… |
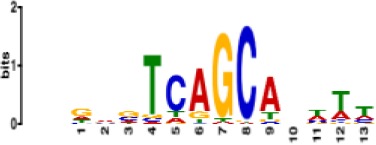
| M2296_1.02 MAFK |
 |
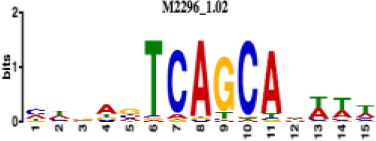 |
629 | M4629_1.02 NFE2, M4572_1.02 MAFF, M4681_1.02 BACH2… |
| M2292_1.02 JUND |
 |
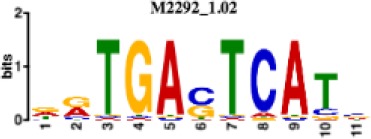 |
571 | M4623_1.02 JUNB, M2278_1.02 FOS, M4619_1.02 FOSL1… |
| M1528_1.02 RFX6 |
 |
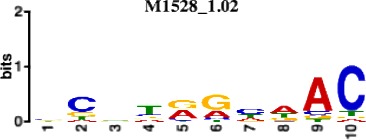 |
556 | M4476_1.02 RFX5, M1529_1.02 RFX7, M5777_1.02 RFX4… |
| M4640_1.02 ZBTB7A |
 |
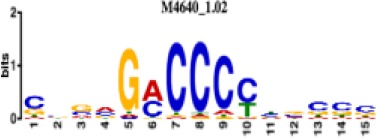 |
530 | M6539_1.02 ZBTB7B, M6552_1.02 ZNF148, M6422_1.02 PLAGL1… |
| M1970_1.02 NFIC |
 |
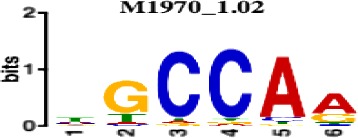 |
484 | M5664_1.02 NFIX, M5660_1.02 NFIA, M5662_1.02 NFIB |
| M2277_1.02 FLI1 |
 |
 |
442 | M6222_1.02 ETV4, M2275_1.02 ELF1, M5398_1.02 ERF… |
| M6281_1.02 HNF1A |
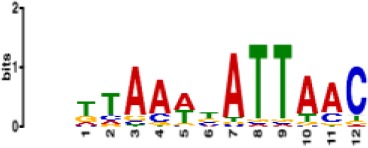 |
 |
418 | M6282_1.02 HNF1B, M6546_1.02 ZFHX3 |
