Figure 3. Two distinct conformations of CK1ε revealed by computational prediction and limited trypsin digestion.
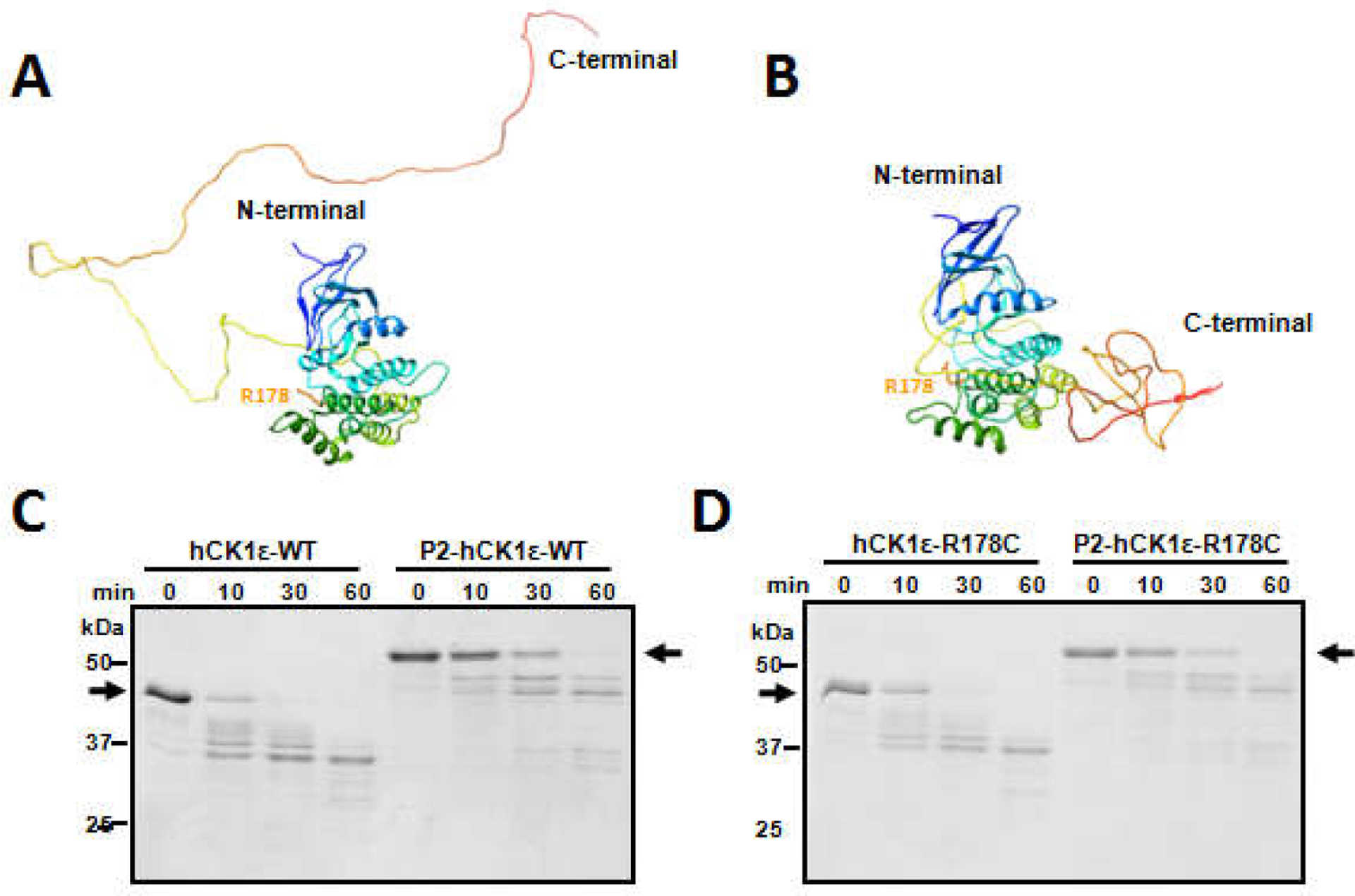
METATASSER was used to predict 3-D structures of CK1ε de novo. Two forms were extracted. In one form, the CTD domain of CK1ε is unstructured and extends away from the N-terminal catalytic domain (A). In the other form, the CTD domain is structured and present immediately adjacent to the catalytic domain (B). Limited trypsin digestion was applied to the differently phosphorylated forms of CK1ε. (C) Non-phosphorylated native CK1ε was digested faster than hyper-phosphorylated native CK1ε. Black arrows indicate the positions of the non-phosphorylatedand hyper-phosphorylatedCK1ε before trypsin digestion. (D) Similar digestion kinetics occur withCK1ε-R178C, indicating that CK1ε-WT and CK1ε-R178C have similar conformations in solution. Data are representativeof two or more independent experiments.
