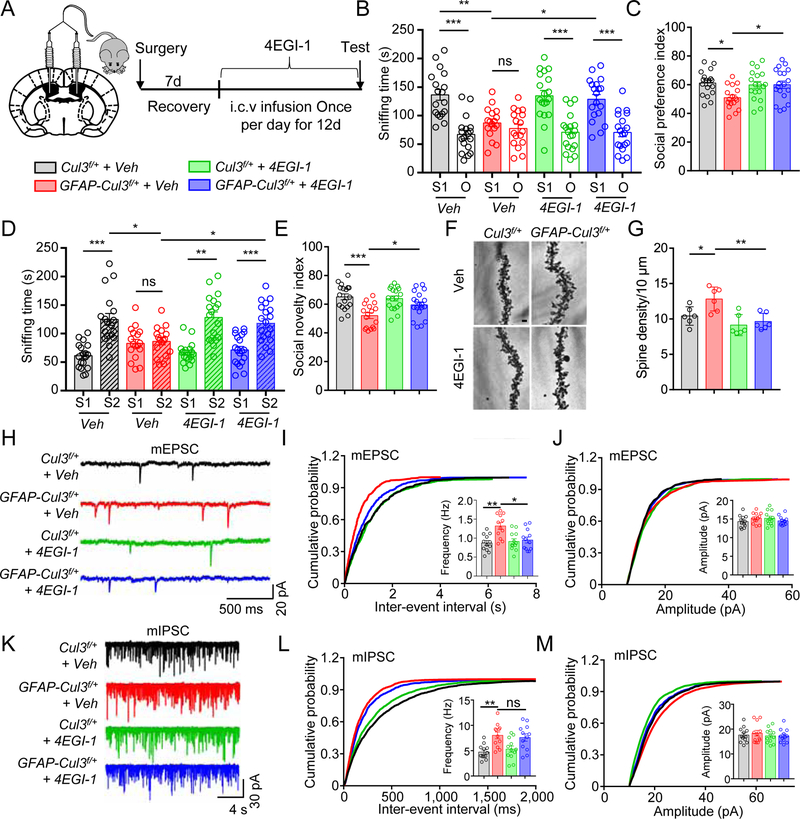
Figure 5. 4EGI-1 ameliorated synaptic and social deficits in CUL3 deficient mice.
(A) Schematic diagram 4EGI-1 i.c.v., bilateral infusion.
(B) 4EGI-1 attenuated social preference deficits in GFAP-Cul3f/+ mice. n = 18 mice for Veh or 4EGI-1 treated Cul3f/+; n = 17 mice for Veh-treated GFAP-Cul3f/+; n = 19 mice for 4EGI-1-treated GFAP-Cul3f/+; Veh-treated Cul3f/+ S1 (136 ± 9.5 s) vs Veh-treated Cul3f/+ O (66.8 ± 7.3 s), p < 0.001; Veh-treated GFAP-Cul3f/+ S1 (87.2 ± 6.6 s) vs Veh-treated GFAP-Cul3f/+ O (77.4 ± 7.6 s), p > 0.999; 4EGI-1-treated Cul3f/+ S1 (135 ± 9.8 s) vs 4EGI-1-treated Cul3f/+ O (67.6 ± 7.0 s), p < 0.001; 4EGI-1-treated GFAP-Cul3f/+ S1 (129 ± 8.5 s) vs 4EGI-1-treated GFAP-Cul3f/+ O (70.6 ± 7.6 s), p < 0.001; Veh-treated Cul3f/+ S1 vs Veh-treated GFAP-Cul3f/+ S1, p = 0.0012; Veh-treated GFAP-Cul3f/+ S1 vs 4EGI-1-treated GFAP-Cul3f/+ S1, p = 0.012; F(1, 136) = 80.24; Two-way ANOVA followed by Bonferroni’s post hoc test.
(C) 4EGI-1 restored social preference index in GFAP-Cul3f/+ mice. n = 18 mice for Veh or 4EGI-1 treated Cul3f/+; n = 17 mice for Veh-treated GFAP-Cul3f/+; n = 19 mice for 4EGI-1-treated GFAP-Cul3f/+; Veh-treated Cul3f/+ (61.2 ± 1.9 s) vs Veh-treated GFAP-Cul3f/+ (51.0 ± 1.8 s), p = 0.0125; Veh-treated GFAP-Cul3f/+ vs 4EGI-1-treated GFAP-Cul3f/+ (60.1 ± 2.5 s), p = 0.031; One-way ANOVA followed by Tukey’s post hoc test.
(D) 4EGI-1 attenuated social novelty deficits in GFAP-Cul3f/+ mice. n = 18 mice for Veh or 4EGI-1 treated Cul3f/+; n = 17 mice for Veh-treated GFAP-Cul3f/+; n = 19 mice for 4EGI-1-treated GFAP-Cul3f/+; Veh-treated Cul3f/+ S1 (61.2 ± 5.2 s) vs Veh-treated Cul3f/+ S2 (125 ± 10.3 s), p < 0.001; Veh-treated GFAP-Cul3f/+ S1 (82.9 ± 6.8 s) vs Veh-treated GFAP-Cul3f/+ S2 (86.3 ± 6.1 s), p > 0.999; 4EGI-1-treated Cul3f/+ S1 (66.5 ± 4.3 s) vs 4EGI-1-treated Cul3f/+ S2 (128 ± 9.6 s), p < 0.001; 4EGI-1-treated GFAP-Cul3f/+ S1 (71.4 ± 6.0 s) vs 4EGI-1-treated GFAP-Cul3f/+ S2 (118 ± 7.9 s), p < 0.001; Veh-treated Cul3f/+ S2 vs Veh-treated GFAP-Cul3f/+ S2, p = 0.0135; Veh-treated GFAP-Cul3f/+ S2 vs 4EGI-1-treated GFAP-Cul3f/+ S2, p = 0.0421; F(1, 136) = 59.13; Two-way ANOVA followed by Bonferroni’s post hoc test.
(E) 4EGI-1 restored social novelty index in GFAP-Cul3f/+ mice. n = 18 mice for Veh or 4EGI-1 treated Cul3f/+ group; n = 17 mice for Veh-treated GFAP-Cul3f/+ group; n = 19 mice for 4EGI-1-treated GFAP-Cul3f/+ group; Veh-treated Cul3f/+ (65.1 ± 1.9 s) vs Veh-treated GFAP-Cul3f/+ (52.0 ± 1.8 s), p = 0.0001; Veh-treated GFAP-Cul3f/+ vs 4EGI-1-treated GFAP-Cul3f/+ (59.5 ± 2.0 s), p = 0.046; One-way ANOVA followed by Tukey’s post hoc test.
(F) Representative Golgi staining images of apical dendrites of CA1 pyramidal neurons. Scale bar, 5 µm.
(G) Dendritic spine density of GFAP-Cul3f/+ neurons was reduced in 4EGI-1-treated GFAP-Cul3f/+ neurons and similar to that of Veh-treated Cul3f/+ neurons. n = 5 mice for all groups; Veh-treated Cul3f/+ (10.4 ± 0.8) vs Veh-treated GFAP-Cul3f/+ (12.8 ± 1.1), p = 0.0394; Veh-treated GFAP-Cul3f/+ vs 4EGI-1-treated GFAP-Cul3f/+ (9.6 ± 0.8), p = 0.0049; One-way ANOVA followed by Tukey’s post hoc test.
(H) Representative mEPSC traces.
(I) mEPSC frequency of GFAP-Cul3f/+ neurons was reduced in 4EGI-1-treated GFAP-Cul3f/+ neurons and similar to that of Veh-treated Cul3f/+ neurons. n = 12 neurons, 3 mice per each group; Veh-treated Cul3f/+ (0.87 ± 0.09 Hz) vs Veh-treated GFAP-Cul3f/+ (1.32 ± 0.15 Hz), p = 0.0071; 4EGI-1-treated GFAP-Cul3f/+ (0.95 ± 0.13 Hz) vs Veh-treated GFAP-Cul3f/+, p = 0.0351; Kruskal-Wallis ANOVA followed by Dunn’s post hoc test.
(J) No effect of 4EGI-1 on mEPSC amplitudes. n = 12 neurons, 3 mice per each group; p > 0.9000 for all comparisons; Kruskal-Wallis ANOVA followed by Dunn’s post hoc test.
(K) Representative mIPSC traces.
(L) mIPSC frequency of GFAP-Cul3f/+ neurons remained increased in 4EGI-1-treated GFAP-Cul3f/+ neurons. n = 12 neurons, 3 mice per each group; Veh-treated Cul3f/+ (4.7 ± 0.58 Hz) vs Veh-treated GFAP-Cul3f/+ (8.0 ± 1.0 Hz), p = 0.0090; 4EGI-1-treated GFAP-Cul3f/+ (7.6 ± 1.02 Hz) vs Veh-treated GFAP-Cul3f/+, p > 0.9999; Kruskal-Wallis ANOVA followed by Dunn’s post hoc test.
(M) No effect of 4EGI-1 on mIPSC amplitudes. n = 12 neurons, 3 mice per each group; p > 0.9999 for all comparisons; Kruskal-Wallis ANOVA followed by Dunn’s post hoc test.
Data were shown as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001; ns, no significant difference.